bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0734_orf1
Length=366
Score E
Sequences producing significant alignments: (Bits) Value
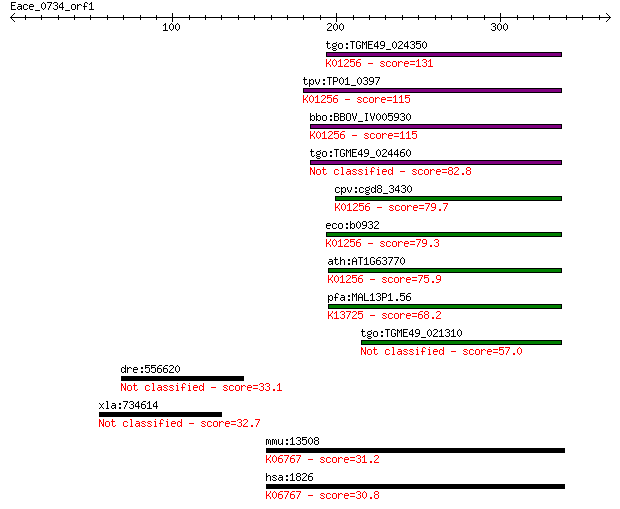
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 131 3e-30
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 115 2e-25
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 115 4e-25
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 82.8 2e-15
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 79.7 2e-14
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 79.3 2e-14
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 75.9 2e-13
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 68.2 5e-11
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 57.0 1e-07
dre:556620 fi40d09; wu:fi40d09 33.1 1.7
xla:734614 rgs3, MGC115680; regulator of G-protein signaling 3 32.7
mmu:13508 Dscam, 4932410A21Rik; Down syndrome cell adhesion mo... 31.2 6.7
hsa:1826 DSCAM, CHD2-42, CHD2-52; Down syndrome cell adhesion ... 30.8 8.8
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 131 bits (330), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 66/143 (46%), Positives = 94/143 (65%), Gaps = 0/143 (0%)
Query 194 RPGEKFRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVAGTPPTDLALNGDDLDLA 253
+P EK R +YKP+D+ +D VDL F L + TKV++T+ M R TPPTDL L+G+DL+L
Sbjct 519 QPVEKHRLDYKPTDFLIDFVDLDFDLYDDRTKVTSTLTMHRREQTPPTDLVLDGEDLELE 578
Query 254 SLMVNGKQVKHIGDSNAVAEGEAGYRLGSDGSLIISKAVLPQKAGEPFTLQTEVSIHPKS 313
S+ ++G + A + Y L DG L+I+ +LPQ+A + F ++T V + PK
Sbjct 579 SVELDGNALSMHSTETQKAGDKRVYSLDVDGRLVIAADLLPQEAEKKFKVKTVVYVRPKE 638
Query 314 NLKLKGLYVSGSALVTQCEAEGY 336
NL+L GLY SG+ LVTQCEAEG+
Sbjct 639 NLQLMGLYKSGALLVTQCEAEGF 661
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 115 bits (289), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 64/157 (40%), Positives = 95/157 (60%), Gaps = 8/157 (5%)
Query 180 TRSELPAMTPKQRGRPGEKFRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVAGTP 239
T SE ++T K + E FR +YKP ++ V++V L F LE+T TKV + ++M+R T
Sbjct 137 TESETSSVTLKPVKKFVEIFRKDYKPPEFDVENVYLEFDLEDTETKVRSKLLMRRRPNTL 196
Query 240 PTDLALNGDDLDLASLMVNGKQVKHIGDSNAVAEGEAGYRLGSDGSLIISKAVLPQKAGE 299
P +L LNGDDL + V+G ++K++ S GY L DG++ + A LP+
Sbjct 197 PGNLTLNGDDLTCNFVAVDGVELKNVPVS--------GYTLDVDGNMTVPSAFLPKDDSR 248
Query 300 PFTLQTEVSIHPKSNLKLKGLYVSGSALVTQCEAEGY 336
FT++T V+I+PK+N KL GLY S +A TQCE G+
Sbjct 249 TFTVETHVTINPKNNTKLVGLYKSSAAFCTQCEPHGF 285
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 115 bits (287), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 64/153 (41%), Positives = 89/153 (58%), Gaps = 8/153 (5%)
Query 184 LPAMTPKQRGRPGEKFRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVAGTPPTDL 243
+ +T K + E FR +Y P+ Y +DSV L F L ET T V A ++M R GT P DL
Sbjct 19 VQTLTLKPAKQHVEIFRKDYTPTIYDIDSVFLDFDLHETATVVKAELLMHRKEGTAPADL 78
Query 244 ALNGDDLDLASLMVNGKQVKHIGDSNAVAEGEAGYRLGSDGSLIISKAVLPQKAGEPFTL 303
L+GD+LD S+ V+GK +++ +GY + DG L I + LP KAGE F +
Sbjct 79 VLHGDELDCRSVSVDGKPLEN--------RPLSGYHIDDDGFLNIPVSFLPSKAGESFRV 130
Query 304 QTEVSIHPKSNLKLKGLYVSGSALVTQCEAEGY 336
TEV I+P +NL+L GLY + TQCE+ G+
Sbjct 131 NTEVVINPTANLQLSGLYKNSQLFTTQCESHGF 163
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 82.8 bits (203), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 57/171 (33%), Positives = 92/171 (53%), Gaps = 19/171 (11%)
Query 184 LPAMTPKQRGRPGEK-------FRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVA 236
L ++ Q R GE+ R +YKP ++ ++ + L F L++ +T V A + + R A
Sbjct 36 LASVITVQVSRAGEEKEEPKAVVRLDYKPPNFYLEDIVLDFNLDDEDTTVEALLTLYRRA 95
Query 237 GTPPTDLALNGDD------LDLA-----SLMVNGKQVKHIGDSNAVAEGEAGYRLGSDGS 285
GT P +L L+GD+ + LA SL G+ D + + +R+ DG+
Sbjct 96 GTEPMNLVLDGDESVILKSISLATTSSPSLRGQGRVTFTELDGQMPTDSQFQFRI-VDGN 154
Query 286 LIISKAVLPQKAGEPFTLQTEVSIHPKSNLKLKGLYVSGSALVTQCEAEGY 336
L++SK +LP++A F ++T VSI+PK+NL L GLY +G VT E G+
Sbjct 155 LMVSKFLLPREAEVRFYIRTLVSINPKANLALFGLYKAGDIFVTLNEPSGF 205
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 79.7 bits (195), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/141 (36%), Positives = 77/141 (54%), Gaps = 12/141 (8%)
Query 199 FRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVAGTPPT-DLALNGDDLDLASLMV 257
+R EYK ++ +D V+L +++ T VS+ I+M+R + DL+L+GD L L S+ +
Sbjct 31 YRKEYKVPNFLIDHVNLDINIKDDVTVVSSVIIMRRNPNSSFRGDLSLDGDCLKLVSVKL 90
Query 258 NGKQVKHIGDSNAVAEGEAGYRL--GSDGSLIISKAVLPQKAGEPFTLQTEVSIHPKSNL 315
NG ++ GY G DG L+IS +LP K + FTL+T V I P N
Sbjct 91 NGVILEK--------SLYKGYFQPDGPDGKLVISSNLLPNK-DQQFTLETVVEIFPDRNT 141
Query 316 KLKGLYVSGSALVTQCEAEGY 336
K GLY S TQCE++G+
Sbjct 142 KNMGLYYSAGVYSTQCESDGF 162
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 79.3 bits (194), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 57/144 (39%), Positives = 76/144 (52%), Gaps = 23/144 (15%)
Query 194 RPGEKFRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVA-GTPPTDLALNGDDLDL 252
+P K+R +Y+ DY + +DL F L+ T V+A V Q V G L LNG+DL L
Sbjct 4 QPQAKYRHDYRAPDYQITDIDLTFDLDAQKTVVTA--VSQAVRHGASDAPLRLNGEDLKL 61
Query 253 ASLMVNGKQVKHIGDSNAVAEGEAGYRLGSDGSLIISKAVLPQKAGEPFTLQTEVSIHPK 312
S+ HI D A E +G+L+IS LP++ FTL+ I P
Sbjct 62 VSV--------HINDEPWTAWKE------EEGALVISN--LPER----FTLKIINEISPA 101
Query 313 SNLKLKGLYVSGSALVTQCEAEGY 336
+N L+GLY SG AL TQCEAEG+
Sbjct 102 ANTALEGLYQSGDALCTQCEAEGF 125
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 75.9 bits (185), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 71/143 (49%), Gaps = 18/143 (12%)
Query 195 PGEKFRDEYKPSDYTVDSVDLRFLLEETNTKVSATI-VMQRVAGTPPTDLALNGDDLDLA 253
P E F Y DY ++VDL F L E T VS+ I V RV G+ L L+G DL L
Sbjct 96 PKEIFLKNYTKPDYYFETVDLSFSLGEEKTIVSSKIKVSPRVKGSSAA-LVLDGHDLKLL 154
Query 254 SLMVNGKQVKHIGDSNAVAEGEAGYRLGSDGSLIISKAVLPQKAGEPFTLQTEVSIHPKS 313
S+ V GK +K E Y+L S + S LP A E F L+ + I+P
Sbjct 155 SVKVEGKLLK-----------EGDYQLDSRHLTLPS---LP--AEESFVLEIDTEIYPHK 198
Query 314 NLKLKGLYVSGSALVTQCEAEGY 336
N L+GLY S TQCEAEG+
Sbjct 199 NTSLEGLYKSSGNFCTQCEAEGF 221
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 68.2 bits (165), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 69/142 (48%), Gaps = 14/142 (9%)
Query 195 PGEKFRDEYKPSDYTVDSVDLRFLLEETNTKVSATIVMQRVAGTPPTDLALNGDDLDLAS 254
P +R +YKPS + +++V L + + T V + + M DL +G L +
Sbjct 196 PKIHYRKDYKPSGFIINNVTLNINIHDNETIVRSVLDMDISKHNVGEDLVFDGVGLKINE 255
Query 255 LMVNGKQVKHIGDSNAVAEGEAGYRLGSDGSLIISKAVLPQKAGEPFTLQTEVSIHPKSN 314
+ +N K++ EGE Y ++ I SK V K F +EV IHP++N
Sbjct 256 ISINNKKL---------VEGEE-YTYDNEFLTIFSKFVPKSK----FAFSSEVIIHPETN 301
Query 315 LKLKGLYVSGSALVTQCEAEGY 336
L GLY S + +V+QCEA G+
Sbjct 302 YALTGLYKSKNIIVSQCEATGF 323
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 57.0 bits (136), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 44/164 (26%), Positives = 66/164 (40%), Gaps = 42/164 (25%)
Query 215 LRFLLEETNTKVSATIVMQRVAGTPPTDLALNGDDL--DLASLMVNGKQVKHIGDSNAVA 272
L+ L N V A + M R G P +L L+GD L + S+ + G+ +
Sbjct 9 LKMPLFMNNEHVKAALTMYRKPGRPVVNLVLDGDGLIAEKVSVSYERENANEAGNGSKKV 68
Query 273 EGEA----------------------------------------GYRLGSDGSLIISKAV 292
EA G + GSL+I K +
Sbjct 69 SVEATQTFPLSSPDAGNQETRQAYLRSENGSSSPSDKEMISPRPGIAVEKSGSLLICKDI 128
Query 293 LPQKAGEPFTLQTEVSIHPKSNLKLKGLYVSGSALVTQCEAEGY 336
LP ++ + F ++T+V I P+ N +L GLYVS LVT EA+G+
Sbjct 129 LPSESEQRFVVKTQVRISPQDNSRLSGLYVSDGVLVTHNEAQGF 172
> dre:556620 fi40d09; wu:fi40d09
Length=2242
Score = 33.1 bits (74), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 38/82 (46%), Gaps = 10/82 (12%)
Query 69 LVEVPTPSLNATGAL-------FSFSLLPRIQAAAAASSLGEEKREEEEELR-DGYLPSA 120
+V + L+A G L FSFS+ P + + SLG +EEE +R + ++
Sbjct 1281 MVAISQKDLDAMGRLRPKRRISFSFSISPMLPKSKTLFSLGSSSSDEEEAVRLSSFTGTS 1340
Query 121 GESEQHADADDPSPHALRGDGE 142
G E +DP P LR D E
Sbjct 1341 GSLEYSISEEDPGP--LRSDSE 1360
> xla:734614 rgs3, MGC115680; regulator of G-protein signaling
3
Length=331
Score = 32.7 bits (73), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 39/83 (46%), Gaps = 8/83 (9%)
Query 55 SLLSPSSPHAWRRGLVEVPTPSLNATGALFSFSLLP----RIQAAAAASSLG----EEKR 106
+ LSP +P W+R + P PS ++T + LP R++ + +A L EE+
Sbjct 15 TCLSPDNPQHWKRVAIRRPQPSTDSTTSSLPVPALPVPEVRVERSFSAGHLACDSDEEEA 74
Query 107 EEEEELRDGYLPSAGESEQHADA 129
+E E LR SE H D+
Sbjct 75 KEAETLRVPLSIWRTRSESHLDS 97
> mmu:13508 Dscam, 4932410A21Rik; Down syndrome cell adhesion
molecule; K06767 Down syndrome cell adhesion molecule
Length=2013
Score = 31.2 bits (69), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 46/187 (24%), Positives = 79/187 (42%), Gaps = 25/187 (13%)
Query 157 AFPNQDTTEPTETAKPRDSDGDTTRSEL--PAMTPKQRGRPGEKFRDEYKPSDYTVDSVD 214
AF N T + ++ K D +G+ T + L P ++ Q K +P ++ S+
Sbjct 552 AFENNGTLKLSDVQKEVD-EGEYTCNVLVQPQLSTSQSVHVTVKVPPFIQPFEFPRFSIG 610
Query 215 LRFLL--EETNTKVSATIVMQRVAGTPPTDLALNGDDLDL-ASLMVNGKQVKHIGDSNAV 271
R + + + TI Q+ P L + D++D +SL ++ + H G+ +
Sbjct 611 QRVFIPCVVVSGDLPITITWQKDGRPIPASLGVTIDNIDFTSSLRISNLSLMHNGNYTCI 670
Query 272 AEGEAGYRLGSDGSLIISKAVLPQKAGEPFTLQTEVSIHPKSNLKLKGLYVSGSALVTQC 331
A EA + LI+ +P K F +Q P+ G+Y G A++ C
Sbjct 671 ARNEAA-AVEHQSQLIVR---VPPK----FVVQ------PRDQ---DGIY--GKAVILNC 711
Query 332 EAEGYIV 338
AEGY V
Sbjct 712 SAEGYPV 718
> hsa:1826 DSCAM, CHD2-42, CHD2-52; Down syndrome cell adhesion
molecule; K06767 Down syndrome cell adhesion molecule
Length=2012
Score = 30.8 bits (68), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 46/187 (24%), Positives = 79/187 (42%), Gaps = 25/187 (13%)
Query 157 AFPNQDTTEPTETAKPRDSDGDTTRSEL--PAMTPKQRGRPGEKFRDEYKPSDYTVDSVD 214
AF N T + ++ K D +G+ T + L P ++ Q K +P ++ S+
Sbjct 552 AFENNGTLKLSDVQKEVD-EGEYTCNVLVQPQLSTSQSVHVTVKVPPFIQPFEFPRFSIG 610
Query 215 LRFLL--EETNTKVSATIVMQRVAGTPPTDLALNGDDLDL-ASLMVNGKQVKHIGDSNAV 271
R + + + TI Q+ P L + D++D +SL ++ + H G+ +
Sbjct 611 QRVFIPCVVVSGDLPITITWQKDGRPIPGSLGVTIDNIDFTSSLRISNLSLMHNGNYTCI 670
Query 272 AEGEAGYRLGSDGSLIISKAVLPQKAGEPFTLQTEVSIHPKSNLKLKGLYVSGSALVTQC 331
A EA + LI+ +P K F +Q P+ G+Y G A++ C
Sbjct 671 ARNEAA-AVEHQSQLIVR---VPPK----FVVQ------PRDQ---DGIY--GKAVILNC 711
Query 332 EAEGYIV 338
AEGY V
Sbjct 712 SAEGYPV 718
Lambda K H
0.312 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 15956348688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40