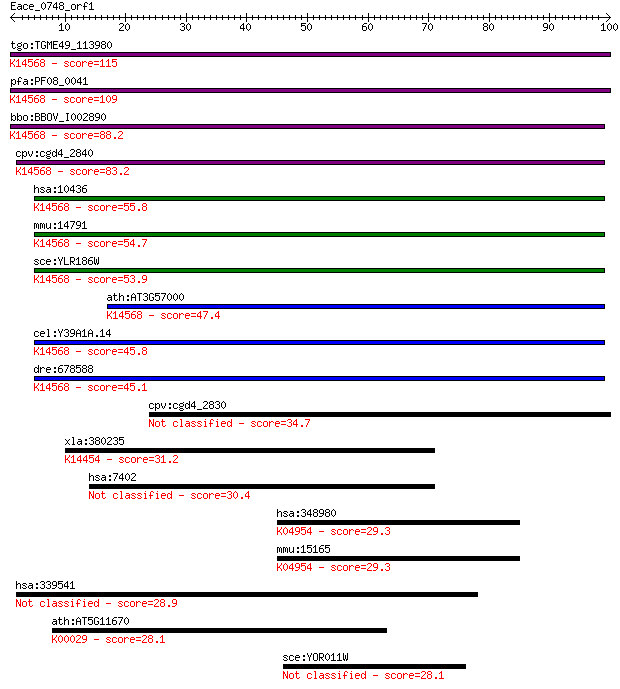
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0748_orf1
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_113980 nucleolar essential protein 1, putative ; K1... 115 3e-26
pfa:PF08_0041 ribosome biogenesis protein nep1 homologue, puta... 109 2e-24
bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain cont... 88.2 5e-18
cpv:cgd4_2840 Mra1/NEP1 like protein, involved in pre-rRNA pro... 83.2 2e-16
hsa:10436 EMG1, C2F, FLJ60792, Grcc2f, NEP1; EMG1 nucleolar pr... 55.8 3e-08
mmu:14791 Emg1, C2f, Grcc2f; EMG1 nucleolar protein homolog (S... 54.7 7e-08
sce:YLR186W EMG1, NEP1; Emg1p; K14568 essential for mitotic gr... 53.9 1e-07
ath:AT3G57000 nucleolar essential protein-related; K14568 esse... 47.4 1e-05
cel:Y39A1A.14 hypothetical protein; K14568 essential for mitot... 45.8 4e-05
dre:678588 MGC136360; zgc:136360; K14568 essential for mitotic... 45.1 5e-05
cpv:cgd4_2830 Mra1/NEP1 like protein, involved in pre-rRNA pro... 34.7 0.082
xla:380235 got1, MGC130786, MGC52828, xr406; glutamic-oxaloace... 31.2 0.89
hsa:7402 UTRN, DMDL, DRP, DRP1, FLJ23678; utrophin 30.4 1.4
hsa:348980 HCN1, BCNG-1, BCNG1, HAC-2; hyperpolarization activ... 29.3 3.1
mmu:15165 Hcn1, Bcng1, C630013B14Rik, HAC2; hyperpolarization-... 29.3 3.4
hsa:339541 C1orf228, MGC33556, NCRNA00082, p40; chromosome 1 o... 28.9 4.3
ath:AT5G11670 ATNADP-ME2 (NADP-malic enzyme 2); malate dehydro... 28.1 6.9
sce:YOR011W AUS1; Aus1p 28.1 7.3
> tgo:TGME49_113980 nucleolar essential protein 1, putative ;
K14568 essential for mitotic growth 1
Length=235
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 53/99 (53%), Positives = 73/99 (73%), Gaps = 0/99 (0%)
Query 1 KNVTLLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHAN 60
KNVTL Q+ KN+ FLP S K+A + GR V L +FVQ+F+ T+ PV+F+VGAVAH++
Sbjct 137 KNVTLAQIVKNEHANFLPPNSVKVALTVSGRSVALSDFVQRFKETDTPVVFVVGAVAHSD 196
Query 61 PTANNELAEECISISPCGLSAAVCCSSLCTEFENLWNIY 99
PT + ++ ISI+ GL+AAVCCSS+C EFE LW+I+
Sbjct 197 PTGECDYVDDKISIAGVGLTAAVCCSSICAEFEALWDIF 235
> pfa:PF08_0041 ribosome biogenesis protein nep1 homologue, putative;
K14568 essential for mitotic growth 1
Length=279
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 48/99 (48%), Positives = 72/99 (72%), Gaps = 0/99 (0%)
Query 1 KNVTLLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHAN 60
KN+ LL++ KND LPI KI S KG+ V+L N+++ +++T PV F +GAVA++N
Sbjct 181 KNIYLLKIIKNDLQNILPINGHKIGLSLKGKKVELNNYIKVYKNTNQPVTFFIGAVAYSN 240
Query 61 PTANNELAEECISISPCGLSAAVCCSSLCTEFENLWNIY 99
PT ++ ++ ISIS LSAA+CCSS+C+EFE+LWN++
Sbjct 241 PTMKLQILDDNISISDFSLSAAMCCSSICSEFEHLWNLF 279
> bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain containing
protein; K14568 essential for mitotic growth 1
Length=188
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 61/98 (62%), Gaps = 0/98 (0%)
Query 1 KNVTLLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHAN 60
++ L++ KN+ + LP GSR+ S GR V L++F QF+ + PV+F +GAV+H +
Sbjct 90 RDAILMRFIKNEIEIALPPGSRRFGLSVGGRQVKLKDFCHQFKAVDYPVVFHIGAVSHTH 149
Query 61 PTANNELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
P E EE +SIS GL+AA C+ +C+EFE L +
Sbjct 150 PKGTVEHVEEVLSISDHGLTAAHVCAKVCSEFEYLLGV 187
> cpv:cgd4_2840 Mra1/NEP1 like protein, involved in pre-rRNA processing,
adjacent genes paralogs ; K14568 essential for mitotic
growth 1
Length=223
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/103 (38%), Positives = 60/103 (58%), Gaps = 6/103 (5%)
Query 2 NVTLLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTEN------PVLFLVGA 55
N +L+++ KND + LP+G +K S G ++ + + +E+ V F+VGA
Sbjct 120 NTSLMRIVKNDIDKILPVGGKKYGLSLNGTQKNIRALINELYSSEDYKIRNSSVTFVVGA 179
Query 56 VAHANPTANNELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
VA+ +P + + EE ISIS LSAA+CCS +C EFE LW I
Sbjct 180 VAYGDPIQSCDFIEEIISISSYPLSAALCCSKICNEFEYLWKI 222
> hsa:10436 EMG1, C2F, FLJ60792, Grcc2f, NEP1; EMG1 nucleolar
protein homolog (S. cerevisiae); K14568 essential for mitotic
growth 1
Length=244
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 53/94 (56%), Gaps = 5/94 (5%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
LL+V KN + P+G K+ S + + + V++ + +P++F+VGA AH +
Sbjct 155 LLKVIKNPVSDHFPVGCMKVGTSFS---IPVVSDVRELVPSSDPIVFVVGAFAHGK--VS 209
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
E E+ +SIS LSAA+ C+ L T FE +W +
Sbjct 210 VEYTEKMVSISNYPLSAALTCAKLTTAFEEVWGV 243
> mmu:14791 Emg1, C2f, Grcc2f; EMG1 nucleolar protein homolog
(S. cerevisiae); K14568 essential for mitotic growth 1
Length=244
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 5/94 (5%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
LL+V KN + P+G KI S V+ + +++ + +PV+F+VGA AH +
Sbjct 155 LLKVIKNPVSDHFPVGCMKIGTSFS---VEDISDIRELVPSSDPVVFVVGAFAHGKVSV- 210
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
E E+ +SIS LSAA+ C+ + T FE +W +
Sbjct 211 -EYTEKMVSISNYPLSAALTCAKVTTAFEEVWGV 243
> sce:YLR186W EMG1, NEP1; Emg1p; K14568 essential for mitotic
growth 1
Length=252
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 49/94 (52%), Gaps = 1/94 (1%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
LL+V KN LP RK+ S + +++++++ E+ +F VGA+A
Sbjct 159 LLKVIKNPITDHLPTKCRKVTLSFDAPVIRVQDYIEKLDDDESICVF-VGAMARGKDNFA 217
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
+E +E + +S LSA+V CS C E+ WNI
Sbjct 218 DEYVDEKVGLSNYPLSASVACSKFCHGAEDAWNI 251
> ath:AT3G57000 nucleolar essential protein-related; K14568 essential
for mitotic growth 1
Length=298
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Query 17 LPIGSRKIAFS-SKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTANNELAEECISIS 75
LP+ S +I FS S + V+++ + + +F+VGA+AH N +E +S+S
Sbjct 217 LPVNSHRIGFSHSSEKLVNMQKHLATVCDDDRDTVFVVGAMAHGKIDCN--YIDEFVSVS 274
Query 76 PCGLSAAVCCSSLCTEFENLWNI 98
LSAA C S +C WNI
Sbjct 275 EYPLSAAYCISRICEALATNWNI 297
> cel:Y39A1A.14 hypothetical protein; K14568 essential for mitotic
growth 1
Length=231
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 48/94 (51%), Gaps = 3/94 (3%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTAN 64
L+ V KN + LP+GSRK+ S + + N + T+ P++ ++G +A +
Sbjct 140 LMSVVKNPVSNHLPVGSRKMLMSFNVPELTMANKLVA-PETDEPLVLIIGGIARGKIVVD 198
Query 65 NELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
+E IS P LSAA+ C+ + + E +W I
Sbjct 199 YNDSETKISNYP--LSAALTCAKVTSGLEEIWGI 230
> dre:678588 MGC136360; zgc:136360; K14568 essential for mitotic
growth 1
Length=238
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 48/95 (50%), Gaps = 7/95 (7%)
Query 5 LLQVTKNDPNLFLPIGSRKIAFSSK-GRHVDLENFVQQFQHTENPVLFLVGAVAHANPTA 63
LL++ KN + LP G + + S K G V V + P ++GA AH
Sbjct 149 LLRLIKNPVSDHLPPGCPRFSTSFKAGDAVCPRTIVPD----DGPAAIVIGAFAHG--AV 202
Query 64 NNELAEECISISPCGLSAAVCCSSLCTEFENLWNI 98
N + E+ +SIS LSAA+ C+ +C+ FE +W +
Sbjct 203 NVDYTEKTVSISNYPLSAALACAKICSAFEEVWGV 237
> cpv:cgd4_2830 Mra1/NEP1 like protein, involved in pre-rRNA processing,
adjacent genes paralogs
Length=216
Score = 34.7 bits (78), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 20/82 (24%), Positives = 38/82 (46%), Gaps = 6/82 (7%)
Query 24 IAFSSKGRHVDLENFVQ-----QFQHTENPVLFLVGAVAHANPTAN-NELAEECISISPC 77
+ S + V L+ F + + ++ + + F++GA A N IS+S
Sbjct 135 VGLSRMAKKVSLQEFCRNEIATKIRNGADNINFVIGASATNNSCGQFTSKFTHYISLSDV 194
Query 78 GLSAAVCCSSLCTEFENLWNIY 99
+ + +CC+ +C+E E L IY
Sbjct 195 SMPSYICCTKICSEMEELLGIY 216
> xla:380235 got1, MGC130786, MGC52828, xr406; glutamic-oxaloacetic
transaminase 1, soluble (aspartate aminotransferase 1);
K14454 aspartate aminotransferase, cytoplasmic [EC:2.6.1.1]
Length=411
Score = 31.2 bits (69), Expect = 0.89, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query 10 KNDPNLFLPIGSRKIA----FSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPTANN 65
+N +F+ G + I + + R +DLE F+Q ++ +FL+ A AH NPT +
Sbjct 140 ENHNAVFMDAGFKDIRAYRYWDAAKRGLDLEGFLQDLENAPEFSIFLLHACAH-NPTGTD 198
Query 66 ELAEE 70
+E
Sbjct 199 PTPDE 203
> hsa:7402 UTRN, DMDL, DRP, DRP1, FLJ23678; utrophin
Length=3433
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 14 NLFLPIGSRKIAFSSKGRHV-----DLENFVQQFQHTENPVLFLVGAVAHANPTANNELA 68
NL I R+ A ++ R V DLENF++ Q E V LV A N ++ LA
Sbjct 2426 NLKQSIADRQNALEAEWRTVQASRRDLENFLKWIQEAETTVNVLVDASHRENALQDSILA 2485
Query 69 EE 70
E
Sbjct 2486 RE 2487
> hsa:348980 HCN1, BCNG-1, BCNG1, HAC-2; hyperpolarization activated
cyclic nucleotide-gated potassium channel 1; K04954 hyperpolarization
activated cyclic nucleotide-gated potassium
channel 1
Length=890
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 11/43 (25%), Positives = 25/43 (58%), Gaps = 3/43 (6%)
Query 45 TENPVLFLVGAVAHAN---PTANNELAEECISISPCGLSAAVC 84
T++P ++ +++H+N P+ + + + +SPC + AVC
Sbjct 662 TQSPPVYTATSLSHSNLHSPSPSTQTPQPSAILSPCSYTTAVC 704
> mmu:15165 Hcn1, Bcng1, C630013B14Rik, HAC2; hyperpolarization-activated,
cyclic nucleotide-gated K+ 1; K04954 hyperpolarization
activated cyclic nucleotide-gated potassium channel
1
Length=910
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 11/43 (25%), Positives = 25/43 (58%), Gaps = 3/43 (6%)
Query 45 TENPVLFLVGAVAHAN---PTANNELAEECISISPCGLSAAVC 84
T++P ++ +++H+N P+ + + + +SPC + AVC
Sbjct 651 TQSPPVYTATSLSHSNLHSPSPSTQTPQPSAILSPCSYTTAVC 693
> hsa:339541 C1orf228, MGC33556, NCRNA00082, p40; chromosome 1
open reading frame 228
Length=440
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 37/82 (45%), Gaps = 16/82 (19%)
Query 2 NVTLLQVTKNDPNLFLPI-----GSRKIA-FSSKGRHVDLENFVQQFQHTENPVLFLVGA 55
+V LLQV N + + G R IA F +K + + T+ V L+ +
Sbjct 125 SVKLLQVIANSGRTYKELICESYGVRSIAEFLAKSKS----------EETQEEVQVLLDS 174
Query 56 VAHANPTANNELAEECISISPC 77
+ H NP N++ + I++ PC
Sbjct 175 LVHGNPKYQNQVYKGLIALLPC 196
> ath:AT5G11670 ATNADP-ME2 (NADP-malic enzyme 2); malate dehydrogenase
(oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/
oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor;
K00029 malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+)
[EC:1.1.1.40]
Length=588
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 8 VTKNDPNLFLPIGSRKIAFSSKGRHVDLENFVQQFQHTENPVLFLVGAVAHANPT 62
+T+ ++L + S+ + SS R L++F Q + H PV L+GAV PT
Sbjct 359 ITETRKKIWL-VDSKGLIVSS--RKESLQHFKQPWAHEHKPVKDLIGAVNAIKPT 410
> sce:YOR011W AUS1; Aus1p
Length=1394
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 10/30 (33%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 46 ENPVLFLVGAVAHANPTANNELAEECISIS 75
+NP F++ AV ++N +A + AEE ++++
Sbjct 984 DNPADFVIDAVGNSNSSAGKDTAEEALTLN 1013
Lambda K H
0.320 0.133 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2046143372
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40