bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0756_orf2
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
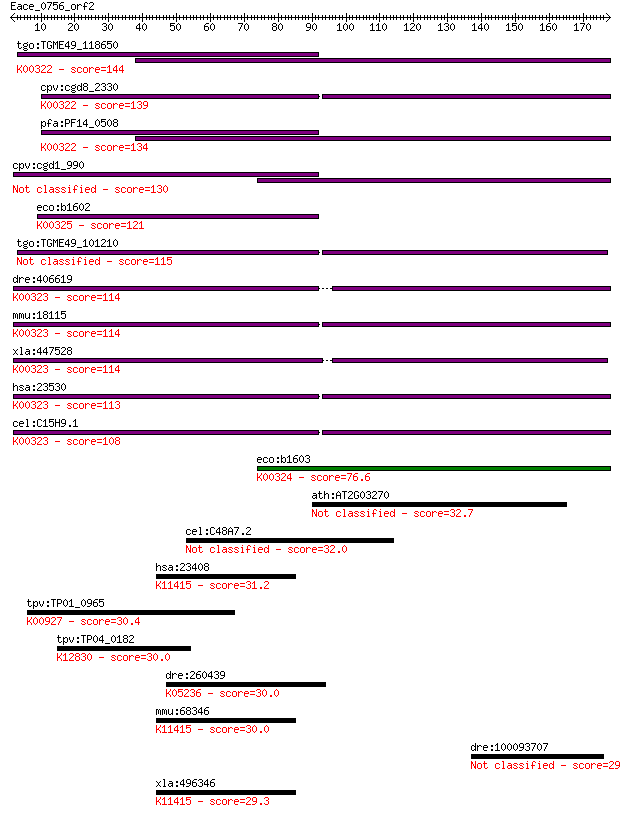
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 144 1e-34
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 139 4e-33
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 134 2e-31
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 130 2e-30
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 121 1e-27
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 115 7e-26
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 114 1e-25
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 114 2e-25
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 114 2e-25
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 113 4e-25
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 108 9e-24
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 76.6 3e-14
ath:AT2G03270 DNA-binding protein, putative 32.7 0.66
cel:C48A7.2 hypothetical protein; K14640 solute carrier family... 32.0 1.1
hsa:23408 SIRT5, FLJ36950, SIR2L5; sirtuin 5; K11415 NAD-depen... 31.2 2.0
tpv:TP01_0965 phosphoglycerate kinase; K00927 phosphoglycerate... 30.4 3.1
tpv:TP04_0182 hypothetical protein; K12830 splicing factor 3B ... 30.0 3.8
dre:260439 copa, cb281, fb13c12, wu:fb13c12; coatomer protein ... 30.0 4.8
mmu:68346 Sirt5, 0610012J09Rik, 1500032M05Rik, AV001953; sirtu... 30.0 4.9
dre:100093707 wu:fj43a03; zgc:165628 29.6 5.9
xla:496346 sirt5; sirtuin 5; K11415 NAD-dependent deacetylase ... 29.3 6.8
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 78/89 (87%), Gaps = 0/89 (0%)
Query 3 QNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGA 62
Q L + G+ V+FGIHPVAGRMPGHMNVLLAEA+V Y+ VKEMSEVN +M YDVV+VVGA
Sbjct 325 QILASRGVEVEFGIHPVAGRMPGHMNVLLAEANVSYRSVKEMSEVNKQMLEYDVVIVVGA 384
Query 63 NDTVNPAALEPGSKISGMPVIEAWKARRV 91
NDTVNPA+LEPG+KI GMPVIE WKA+RV
Sbjct 385 NDTVNPASLEPGTKIYGMPVIEVWKAKRV 413
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 68/141 (48%), Positives = 92/141 (65%), Gaps = 12/141 (8%)
Query 38 YKIVKEMSEVNPEMSSYDVVLVVGANDTVNPA-ALEPGSKISGMPVIEAWKARRVFAHGL 96
Y+ V E P +S + A V+ A L G+ ++G+ I AHGL
Sbjct 636 YRAVLEAFNALPRLSKTSIT----AAGRVDAARVLVLGAGVAGLQAIST-------AHGL 684
Query 97 GAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIK 156
A+VF +DVR+ATREEVESCGG F+ + + EE EV GGYAREMG+AY+ AQ+++++ +
Sbjct 685 QAEVFAYDVRAATREEVESCGGTFLSVELEEEGEVEGGYAREMGEAYEMAQKQMLSRVVP 744
Query 157 HCDVVICTAAIHGKPSPKLIS 177
+ DV+ICTAAIHGKPSPKLIS
Sbjct 745 NVDVIICTAAIHGKPSPKLIS 765
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 139 bits (350), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 64/82 (78%), Positives = 71/82 (86%), Gaps = 0/82 (0%)
Query 10 ITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPA 69
I V+F IHPVAGRMPGHMNVLLAEADVPY IVKEM +VNP M S+DVVLV+GANDTVNP
Sbjct 426 IYVEFAIHPVAGRMPGHMNVLLAEADVPYSIVKEMEDVNPHMESFDVVLVIGANDTVNPL 485
Query 70 ALEPGSKISGMPVIEAWKARRV 91
ALE SKI+GMPVIE W+A +V
Sbjct 486 ALEKDSKINGMPVIEVWRASKV 507
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 57/85 (67%), Gaps = 0/85 (0%)
Query 93 AHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIA 152
A LGA V+ D R+ATREEVES G KF+ + + E+ + GYA+ M Y +AQ +L +
Sbjct 771 AKNLGADVYASDTRTATREEVESLGAKFVTVDIKEDGDSGSGYAKVMSPEYLKAQSKLYS 830
Query 153 NTIKHCDVVICTAAIHGKPSPKLIS 177
I+ CDVVI TA I GKPSPK+I+
Sbjct 831 KMIRSCDVVITTALIPGKPSPKIIT 855
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 134 bits (337), Expect = 2e-31, Method: Composition-based stats.
Identities = 58/82 (70%), Positives = 71/82 (86%), Gaps = 0/82 (0%)
Query 10 ITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPA 69
I V F IHPVAGRMPGH+NVLLAEA++PY IVKEM+E+NP +S D+VLVVGAND VNP+
Sbjct 436 INVTFAIHPVAGRMPGHLNVLLAEANIPYNIVKEMNEINPIISEADIVLVVGANDIVNPS 495
Query 70 ALEPGSKISGMPVIEAWKARRV 91
+L+P SKI GMPVIE WK+++V
Sbjct 496 SLDPSSKIYGMPVIEVWKSKQV 517
Score = 84.0 bits (206), Expect = 3e-16, Method: Composition-based stats.
Identities = 50/141 (35%), Positives = 72/141 (51%), Gaps = 12/141 (8%)
Query 38 YKIVKEMSEVNPEMSSYDVVLVVGANDTVNPA-ALEPGSKISGMPVIEAWKARRVFAHGL 96
Y+ V E + P S + A +NPA G+ ++G+ I + A L
Sbjct 783 YRAVLEAFFILPRFSKSSIT----AAGKINPAKVFVIGAGVAGLQAI-------ITAKSL 831
Query 97 GAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIK 156
GA V+ HD R AT EEV+SCGG FI + E ++L + Y + Q L IK
Sbjct 832 GAIVYSHDSRLATEEEVKSCGGIFIRIPTSERGDILNMSTDMNNEEYIKVQSNLFKKIIK 891
Query 157 HCDVVICTAAIHGKPSPKLIS 177
CD++IC+A+I GK SPKL++
Sbjct 892 KCDILICSASIPGKTSPKLVT 912
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 130 bits (327), Expect = 2e-30, Method: Composition-based stats.
Identities = 62/90 (68%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 2 IQNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 61
I+ L + V+ GIHPVAGRMPGHMNVLLAE+ VP +IVKEM VN M YD+VLVVG
Sbjct 381 IKELESRSTIVEIGIHPVAGRMPGHMNVLLAESGVPSRIVKEMDAVNNCMHEYDLVLVVG 440
Query 62 ANDTVNPAALEPGSKISGMPVIEAWKARRV 91
AND VNPAAL+P SKISGMPVI WKA++V
Sbjct 441 ANDIVNPAALDPQSKISGMPVINVWKAKQV 470
Score = 50.1 bits (118), Expect = 4e-06, Method: Composition-based stats.
Identities = 33/104 (31%), Positives = 53/104 (50%), Gaps = 15/104 (14%)
Query 74 GSKISGMPVIEAWKARRVFAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLG 133
G+ ++G+ I K+ +G +VF D RS ++EE ESCG +FI ++ E E L
Sbjct 772 GAGVAGLQAIATAKS-------MGTKVFAMDSRSTSKEEAESCGARFI--QVPSEGESL- 821
Query 134 GYAREMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLIS 177
+ + QR+LI + D+VI +A G+ P LI+
Sbjct 822 -----RKEEILKKQRDLIEKYLCISDIVITSACKPGEECPILIT 860
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 58/84 (69%), Positives = 67/84 (79%), Gaps = 1/84 (1%)
Query 9 GITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNP 68
GI V FGIHPVAGR+PGHMNVLLAEA VPY IV EM E+N + + D VLV+GANDTVNP
Sbjct 337 GINVRFGIHPVAGRLPGHMNVLLAEAKVPYDIVLEMDEINDDFADTDTVLVIGANDTVNP 396
Query 69 AAL-EPGSKISGMPVIEAWKARRV 91
AA +P S I+GMPV+E WKA+ V
Sbjct 397 AAQDDPKSPIAGMPVLEVWKAQNV 420
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 115 bits (288), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 55/90 (61%), Positives = 65/90 (72%), Gaps = 1/90 (1%)
Query 3 QNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGA 62
Q L G V F IHPVAGR+PGHMNVLLAEA+VPY IV M E+N + D V+++GA
Sbjct 316 QQLRERGTQVRFAIHPVAGRLPGHMNVLLAEANVPYDIVLSMDEINEDFPQTDTVIIIGA 375
Query 63 NDTVNPAALE-PGSKISGMPVIEAWKARRV 91
NDTVNPAA PG I GMPV+E WKA++V
Sbjct 376 NDTVNPAAQNAPGCPIYGMPVLEVWKAKKV 405
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 54/88 (61%), Gaps = 4/88 (4%)
Query 93 AHGLGAQVFGHDVRSATREEVESCGGKFIGLRM---GEEAEVL-GGYAREMGDAYQRAQR 148
AH LGA V G DVR +E+VES GG+F+ + GE GGYA+ M + + R +
Sbjct 781 AHRLGADVRGFDVRLECKEQVESMGGEFLAMTFEGPGESGRATAGGYAKPMSEEFLRKEM 840
Query 149 ELIANTIKHCDVVICTAAIHGKPSPKLI 176
EL A + D++I TA++ G+P+PKLI
Sbjct 841 ELFAEQCREIDILITTASLPGRPAPKLI 868
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 55/91 (60%), Positives = 70/91 (76%), Gaps = 1/91 (1%)
Query 2 IQNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 61
+++L + G V FGIHPVAGRMPG +NVLLAEA VPY +V EM E+N + D+VLV+G
Sbjct 944 VKSLTDQGKKVRFGIHPVAGRMPGQLNVLLAEAGVPYDVVLEMDEINEDFPETDLVLVIG 1003
Query 62 ANDTVNPAALE-PGSKISGMPVIEAWKARRV 91
ANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 1004 ANDTVNSAAQEDPNSIIAGMPVLEVWKSKQV 1034
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 47/82 (57%), Gaps = 0/82 (0%)
Query 96 LGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTI 155
+GA V G D R+A E+ +S G + + + + E E GGYA+EM + A+ +L A
Sbjct 245 MGAIVRGFDTRAAALEQFKSLGAEPLEVDIKESGEGQGGYAKEMSKEFIEAEMKLFAKQC 304
Query 156 KHCDVVICTAAIHGKPSPKLIS 177
D++I TA I G+ +P LI+
Sbjct 305 LDVDIIITTALIPGRKAPVLIT 326
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/91 (61%), Positives = 68/91 (74%), Gaps = 1/91 (1%)
Query 2 IQNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 61
++ L G V FGIHPVAGRMPG +NVLLAEA VPY IV EM E+N + D+VLV+G
Sbjct 948 VKMLTEQGKKVRFGIHPVAGRMPGQLNVLLAEAGVPYDIVLEMDEINSDFPDTDLVLVIG 1007
Query 62 ANDTVNPAALE-PGSKISGMPVIEAWKARRV 91
ANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 1008 ANDTVNSAAQEDPNSIIAGMPVLEVWKSKQV 1038
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 48/85 (56%), Gaps = 0/85 (0%)
Query 93 AHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIA 152
A +GA V G D R+A E+ +S G + + + + E E GGYA+EM + A+ +L A
Sbjct 246 AKSMGAVVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFA 305
Query 153 NTIKHCDVVICTAAIHGKPSPKLIS 177
K D++I TA I GK +P L S
Sbjct 306 QQCKEVDILISTALIPGKKAPVLFS 330
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/92 (60%), Positives = 68/92 (73%), Gaps = 1/92 (1%)
Query 2 IQNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 61
++ L G V FGIHPVAGRMPG +NVLLAEA VPY IV EM E+N + D+VLV+G
Sbjct 948 VKILKEAGKNVRFGIHPVAGRMPGQLNVLLAEAGVPYDIVLEMDEINEDFPETDLVLVIG 1007
Query 62 ANDTVNPAALE-PGSKISGMPVIEAWKARRVF 92
ANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 1008 ANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVI 1039
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 46/81 (56%), Gaps = 0/81 (0%)
Query 96 LGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTI 155
+GA V G D R+A E+ +S G + + + + E E GGYA+EM + A+ +L A
Sbjct 249 MGAIVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFAKQC 308
Query 156 KHCDVVICTAAIHGKPSPKLI 176
+ D+++ TA I GK +P L
Sbjct 309 QDVDIIVTTALIPGKTAPILF 329
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 56/91 (61%), Positives = 68/91 (74%), Gaps = 1/91 (1%)
Query 2 IQNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 61
++ L G V FGIHPVAGRMPG +NVLLAEA VPY IV EM E+N + D+VLV+G
Sbjct 948 VKMLTEQGKKVRFGIHPVAGRMPGQLNVLLAEAGVPYDIVLEMDEINHDFPDTDLVLVIG 1007
Query 62 ANDTVNPAALE-PGSKISGMPVIEAWKARRV 91
ANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 1008 ANDTVNSAAQEDPNSIIAGMPVLEVWKSKQV 1038
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 48/85 (56%), Gaps = 0/85 (0%)
Query 93 AHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIA 152
A +GA V G D R+A E+ +S G + + + + E E GGYA+EM + A+ +L A
Sbjct 246 AKSMGAIVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFA 305
Query 153 NTIKHCDVVICTAAIHGKPSPKLIS 177
K D++I TA I GK +P L +
Sbjct 306 QQCKEVDILISTALIPGKKAPVLFN 330
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 108 bits (270), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 50/91 (54%), Positives = 66/91 (72%), Gaps = 1/91 (1%)
Query 2 IQNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 61
++ L G+ V F IHPVAGRMPG +NVLLAEA VPY IV+EM E+N + DV LV+G
Sbjct 904 VKELQQRGVRVRFAIHPVAGRMPGQLNVLLAEAGVPYDIVEEMEEINEDFKETDVALVIG 963
Query 62 ANDTVNPAAL-EPGSKISGMPVIEAWKARRV 91
+NDT+N AA +P S I+GMPV+ W +++V
Sbjct 964 SNDTINSAAEDDPNSSIAGMPVLRVWNSKQV 994
Score = 73.2 bits (178), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 36/85 (42%), Positives = 54/85 (63%), Gaps = 0/85 (0%)
Query 93 AHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIA 152
+ G+GA V G D R+A +E VES G +F+ + + E+ E GGYA+EM + A+ +L A
Sbjct 212 SRGMGAVVRGFDTRAAVKEHVESLGAQFLTVNVKEDGEGGGGYAKEMSKEFIDAEMKLFA 271
Query 153 NTIKHCDVVICTAAIHGKPSPKLIS 177
+ K D++I TA I GK +P LI+
Sbjct 272 DQCKDVDIIITTALIPGKKAPILIT 296
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/104 (40%), Positives = 61/104 (58%), Gaps = 7/104 (6%)
Query 74 GSKISGMPVIEAWKARRVFAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLG 133
G+ ++G+ I A A+ LGA V D R +E+V+S G +F+ L EEA
Sbjct 172 GAGVAGLAAIGA-------ANSLGAIVRAFDTRPEVKEQVQSMGAEFLELDFKEEAGSGD 224
Query 134 GYAREMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLIS 177
GYA+ M DA+ +A+ EL A K D+++ TA I GKP+PKLI+
Sbjct 225 GYAKVMSDAFIKAEMELFAAQAKEVDIIVTTALIPGKPAPKLIT 268
> ath:AT2G03270 DNA-binding protein, putative
Length=639
Score = 32.7 bits (73), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 39/78 (50%), Gaps = 3/78 (3%)
Query 90 RVFAHGLGAQVFGHD---VRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRA 146
+V L AQV D + + R+E+++ GK + + ++ R +G ++
Sbjct 272 QVLDSALDAQVLKGDNSGLANDIRKEMKALNGKLLKAKDKNTRRLIQKELRTLGKEERKR 331
Query 147 QRELIANTIKHCDVVICT 164
Q+ +++ IK+ DV++ T
Sbjct 332 QQLAVSDVIKNADVILTT 349
> cel:C48A7.2 hypothetical protein; K14640 solute carrier family
20 (sodium-dependent phosphate transporter)
Length=530
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Query 53 SYDVVLVVGANDTVNPAALEPGSKISGMPVIEAWKARRVFAHGLGAQVFGHDVRSATREE 112
++ + +GANDT N GSK+ + + +A+ +F LGA + GH V R+
Sbjct 16 AFILAFAIGANDTANSFGTSVGSKV--LTLHQAYVLASIF-ETLGACLLGHQVTDTMRKG 72
Query 113 V 113
V
Sbjct 73 V 73
> hsa:23408 SIRT5, FLJ36950, SIR2L5; sirtuin 5; K11415 NAD-dependent
deacetylase sirtuin 5 [EC:3.5.1.-]
Length=292
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 44 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 84
+ EV+ E++ D+ LVVG + V PAA+ P G+PV E
Sbjct 214 LEEVDRELAHCDLCLVVGTSSVVYPAAMFAPQVAARGVPVAE 255
> tpv:TP01_0965 phosphoglycerate kinase; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=415
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/63 (22%), Positives = 35/63 (55%), Gaps = 2/63 (3%)
Query 6 MNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYK--IVKEMSEVNPEMSSYDVVLVVGAN 63
M+C ++ G++ VA ++PG ++ + +VP + ++K+++ V + + +L G +
Sbjct 1 MDCLLSSKLGLNDVADKLPGKRVLMRVDYNVPMRDGVIKDLTRVKATIPTIKFLLENGVH 60
Query 64 DTV 66
V
Sbjct 61 SVV 63
> tpv:TP04_0182 hypothetical protein; K12830 splicing factor 3B
subunit 3
Length=1231
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 4/43 (9%)
Query 15 GIHPVA--GRMPGHMNVLL--AEADVPYKIVKEMSEVNPEMSS 53
G++P G +PG +NVLL E+D +++ E+N EM+S
Sbjct 817 GVNPAVPQGPVPGQLNVLLLVVESDYNSYNTEQVEEINKEMTS 859
> dre:260439 copa, cb281, fb13c12, wu:fb13c12; coatomer protein
complex, subunit alpha; K05236 coatomer protein complex, subunit
alpha (xenin)
Length=1224
Score = 30.0 bits (66), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query 47 VNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIEAWKARRVFA 93
N E S+YD+ + +D+ NP A E G + SG+ + W AR FA
Sbjct 384 TNLENSTYDLYSIPRESDSQNPDAPE-GKRSSGLTAV--WVARNRFA 427
> mmu:68346 Sirt5, 0610012J09Rik, 1500032M05Rik, AV001953; sirtuin
5 (silent mating type information regulation 2 homolog)
5 (S. cerevisiae); K11415 NAD-dependent deacetylase sirtuin
5 [EC:3.5.1.-]
Length=310
Score = 30.0 bits (66), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 44 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 84
+ EV+ E++ D+ LVVG + V PAA+ P G+PV E
Sbjct 232 LEEVDRELALCDLCLVVGTSSVVYPAAMFAPQVASRGVPVAE 273
> dre:100093707 wu:fj43a03; zgc:165628
Length=208
Score = 29.6 bits (65), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 137 REMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKL 175
R+ G+ +QRE+I +H D + A +H P P L
Sbjct 4 RKQGNPQHLSQREIITPEAEHVDAGLAVADLHSHPHPLL 42
> xla:496346 sirt5; sirtuin 5; K11415 NAD-dependent deacetylase
sirtuin 5 [EC:3.5.1.-]
Length=309
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Query 44 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 84
+ EV E+ + D+ +VVG + V PAA+ P G+PV E
Sbjct 231 LGEVEKELETCDLCVVVGTSSVVYPAAMFAPQVAARGVPVAE 272
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40