bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0767_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
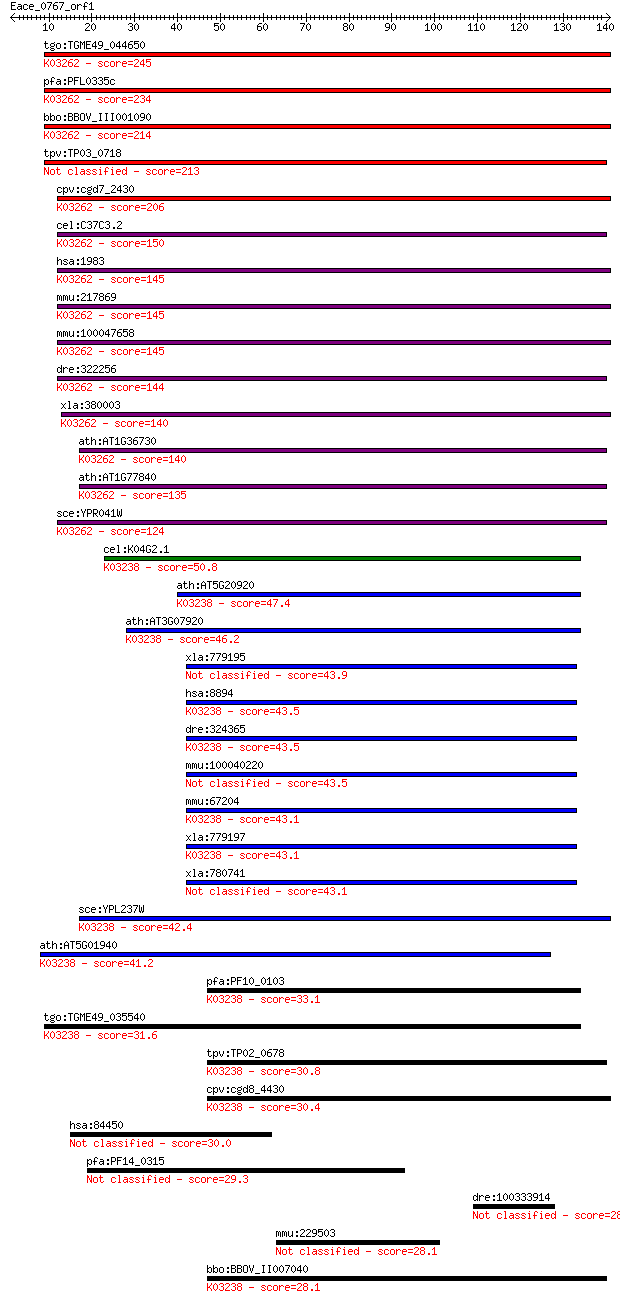
tgo:TGME49_044650 eukaryotic translation initiation factor 5, ... 245 3e-65
pfa:PFL0335c eukaryotic translation initiation factor 5, putat... 234 7e-62
bbo:BBOV_III001090 17.m07123; eukaryotic translation initiatio... 214 8e-56
tpv:TP03_0718 eukaryotic translation initiation factor 5 213 2e-55
cpv:cgd7_2430 translation initiation factor eIF-5; Tif5p, ZnR+... 206 2e-53
cel:C37C3.2 hypothetical protein; K03262 translation initiatio... 150 1e-36
hsa:1983 EIF5, EIF-5A; eukaryotic translation initiation facto... 145 5e-35
mmu:217869 Eif5, 2810011H21Rik, D12Ertd549e, MGC36374, MGC3650... 145 6e-35
mmu:100047658 eukaryotic translation initiation factor 5-like;... 145 6e-35
dre:322256 eif5, fb37c12, fb54h04, wu:fb37c12, wu:fb54h04, zgc... 144 9e-35
xla:380003 eif5, MGC52788; eukaryotic translation initiation f... 140 1e-33
ath:AT1G36730 eukaryotic translation initiation factor 5, puta... 140 1e-33
ath:AT1G77840 eukaryotic translation initiation factor 5, puta... 135 5e-32
sce:YPR041W TIF5, SUI5; Tif5p; K03262 translation initiation f... 124 1e-28
cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiatio... 50.8 1e-06
ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238... 47.4 2e-05
ath:AT3G07920 translation initiation factor; K03238 translatio... 46.2 3e-05
xla:779195 hypothetical protein MGC130708 43.9 2e-04
hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC850... 43.5 2e-04
dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryot... 43.5 2e-04
mmu:100040220 Gm9892; predicted gene 9892 43.5
mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381, AA... 43.1 3e-04
xla:779197 eif2s2, MGC130959; eukaryotic translation initiatio... 43.1 3e-04
xla:780741 eukaryote initiation factor 2 beta 43.1 3e-04
sce:YPL237W SUI3; Beta subunit of the translation initiation f... 42.4 5e-04
ath:AT5G01940 eukaryotic translation initiation factor 2B fami... 41.2 0.001
pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,... 33.1 0.27
tgo:TGME49_035540 translation initiation factor 2 beta, putati... 31.6 0.74
tpv:TP02_0678 eukaryotic translation initiation factor 2 subun... 30.8 1.4
cpv:cgd8_4430 translation initiation factor if-2 betam beta su... 30.4 1.7
hsa:84450 ZNF512, FLJ51176, KIAA1805, MGC111046; zinc finger p... 30.0 2.5
pfa:PF14_0315 conserved Plasmodium membrane protein, unknown f... 29.3 4.3
dre:100333914 C-C motif chemokine 21-like 28.5 6.4
mmu:229503 Rrnad1, MGC38548; ribosomal RNA adenine dimethylase... 28.1 8.3
bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation... 28.1 8.6
> tgo:TGME49_044650 eukaryotic translation initiation factor 5,
putative ; K03262 translation initiation factor 5
Length=414
Score = 245 bits (626), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 111/132 (84%), Positives = 121/132 (91%), Gaps = 0/132 (0%)
Query 9 MQYVNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 68
M VNIPRD++DPNYRYKMPKL+SKIEGRGNGI+TN++NMGEIARALKRPPMYPTKFFGC
Sbjct 1 MALVNIPRDRDDPNYRYKMPKLVSKIEGRGNGIKTNIFNMGEIARALKRPPMYPTKFFGC 60
Query 69 ELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCK 128
ELGAM KFEE EEKAL+NGAH E DLV ILDKFIQMYVLC GCELPEID+ VKKG+L CK
Sbjct 61 ELGAMAKFEEAEEKALVNGAHKESDLVNILDKFIQMYVLCPGCELPEIDLIVKKGLLTCK 120
Query 129 CNACGYSGTLDN 140
CNACGY G+LDN
Sbjct 121 CNACGYQGSLDN 132
> pfa:PFL0335c eukaryotic translation initiation factor 5, putative;
K03262 translation initiation factor 5
Length=565
Score = 234 bits (597), Expect = 7e-62, Method: Compositional matrix adjust.
Identities = 107/132 (81%), Positives = 117/132 (88%), Gaps = 0/132 (0%)
Query 9 MQYVNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 68
M YVNIPRD+ DPNYRYKMPKL+SKIEGRGNGIRTN+ NMGEIAR+LKRPPMYPTKFFGC
Sbjct 1 MSYVNIPRDRNDPNYRYKMPKLISKIEGRGNGIRTNISNMGEIARSLKRPPMYPTKFFGC 60
Query 69 ELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCK 128
ELG MVKFEENEEKA++NGAH E+DLV ILDKFI+MYVLC C LPE DI VKKG+L CK
Sbjct 61 ELGTMVKFEENEEKAIVNGAHKEKDLVNILDKFIEMYVLCPHCLLPETDIVVKKGILICK 120
Query 129 CNACGYSGTLDN 140
CNACG G L+N
Sbjct 121 CNACGNIGELNN 132
> bbo:BBOV_III001090 17.m07123; eukaryotic translation initiation
factor 5; K03262 translation initiation factor 5
Length=379
Score = 214 bits (544), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 100/132 (75%), Positives = 111/132 (84%), Gaps = 0/132 (0%)
Query 9 MQYVNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 68
M VNIPR EDPNYRYKMPKL SKIEGRGNGI+TN+ NMG+IARALKRPP Y TKFFGC
Sbjct 1 MSLVNIPRYCEDPNYRYKMPKLQSKIEGRGNGIKTNISNMGDIARALKRPPTYATKFFGC 60
Query 69 ELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCK 128
ELGAM KFEE+EEKALINGAH+E+ L ILDKFI+MYVLC C LPEI++ VK+G L +
Sbjct 61 ELGAMSKFEESEEKALINGAHTERALTQILDKFIEMYVLCAECHLPEIEVLVKRGALCSR 120
Query 129 CNACGYSGTLDN 140
CNACGY G LDN
Sbjct 121 CNACGYKGVLDN 132
> tpv:TP03_0718 eukaryotic translation initiation factor 5
Length=385
Score = 213 bits (542), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 93/131 (70%), Positives = 112/131 (85%), Gaps = 0/131 (0%)
Query 9 MQYVNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 68
M YVNIPR ++DPNYRYKMP++ S+IEGRGNG +TN+ NMG+IARALKRPP Y TKFFGC
Sbjct 1 MSYVNIPRYRDDPNYRYKMPRIQSRIEGRGNGTKTNISNMGDIARALKRPPTYATKFFGC 60
Query 69 ELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCK 128
ELGAM KFEE+EEKALINGAH++ L +LDKFI++YVLC C+LPEI++ VK+G L C
Sbjct 61 ELGAMSKFEESEEKALINGAHTDTTLAGVLDKFIELYVLCQNCQLPEIELFVKRGELLCS 120
Query 129 CNACGYSGTLD 139
CNACG+ GTLD
Sbjct 121 CNACGHKGTLD 131
> cpv:cgd7_2430 translation initiation factor eIF-5; Tif5p, ZnR+W2
domains ; K03262 translation initiation factor 5
Length=460
Score = 206 bits (524), Expect = 2e-53, Method: Composition-based stats.
Identities = 92/129 (71%), Positives = 108/129 (83%), Gaps = 0/129 (0%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
+NIP +DPNYRYKMPKL+SKIEGRGNGIRT + NM +IAR+LKRPP YPTKFFGCELG
Sbjct 2 LNIPSTVDDPNYRYKMPKLVSKIEGRGNGIRTKIVNMADIARSLKRPPAYPTKFFGCELG 61
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCKCNA 131
A+ K+E+ EEK+++NGAH + DL ++DKFIQMYVLC CELPEIDI VKK + CKCNA
Sbjct 62 ALSKWEDKEEKSIVNGAHQQNDLQRLMDKFIQMYVLCPNCELPEIDIIVKKERVSCKCNA 121
Query 132 CGYSGTLDN 140
CGY G LDN
Sbjct 122 CGYIGALDN 130
> cel:C37C3.2 hypothetical protein; K03262 translation initiation
factor 5
Length=413
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 69/128 (53%), Positives = 89/128 (69%), Gaps = 0/128 (0%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
+N+ R DP YRYKMPKL +K+EG+GNGI+T + NM EIA+AL+RPPMYPTK+FGCELG
Sbjct 3 LNVNRAVADPFYRYKMPKLSAKVEGKGNGIKTVISNMSEIAKALERPPMYPTKYFGCELG 62
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCKCNA 131
A F+ E+ ++NG H L ILD FI+ +VLC CE PE + V+K + KC A
Sbjct 63 AQTNFDAKNERYIVNGEHDANKLQDILDGFIKKFVLCKSCENPETQLFVRKNNIKSKCKA 122
Query 132 CGYSGTLD 139
CG S +D
Sbjct 123 CGCSFDID 130
> hsa:1983 EIF5, EIF-5A; eukaryotic translation initiation factor
5; K03262 translation initiation factor 5
Length=431
Score = 145 bits (365), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 90/131 (68%), Gaps = 2/131 (1%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
VN+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELG
Sbjct 3 VNVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELG 62
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITV--KKGVLFCKC 129
A +F+ ++ ++NG+H L +LD FI+ +VLC CE PE D+ V KK + C
Sbjct 63 AQTQFDVKNDRYIVNGSHEANKLQDMLDGFIKKFVLCPECENPETDLHVNPKKQTIGNSC 122
Query 130 NACGYSGTLDN 140
ACGY G LD
Sbjct 123 KACGYRGMLDT 133
> mmu:217869 Eif5, 2810011H21Rik, D12Ertd549e, MGC36374, MGC36509;
eukaryotic translation initiation factor 5; K03262 translation
initiation factor 5
Length=429
Score = 145 bits (365), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 90/131 (68%), Gaps = 2/131 (1%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
VN+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELG
Sbjct 3 VNVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELG 62
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITV--KKGVLFCKC 129
A +F+ ++ ++NG+H L +LD FI+ +VLC CE PE D+ V KK + C
Sbjct 63 AQTQFDVKNDRYIVNGSHEANKLQDMLDGFIKKFVLCPECENPETDLHVNPKKQTIGNSC 122
Query 130 NACGYSGTLDN 140
ACGY G LD
Sbjct 123 KACGYRGMLDT 133
> mmu:100047658 eukaryotic translation initiation factor 5-like;
K03262 translation initiation factor 5
Length=429
Score = 145 bits (365), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 90/131 (68%), Gaps = 2/131 (1%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
VN+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELG
Sbjct 3 VNVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELG 62
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITV--KKGVLFCKC 129
A +F+ ++ ++NG+H L +LD FI+ +VLC CE PE D+ V KK + C
Sbjct 63 AQTQFDVKNDRYIVNGSHEANKLQDMLDGFIKKFVLCPECENPETDLHVNPKKQTIGNSC 122
Query 130 NACGYSGTLDN 140
ACGY G LD
Sbjct 123 KACGYRGMLDT 133
> dre:322256 eif5, fb37c12, fb54h04, wu:fb37c12, wu:fb54h04, zgc:56606,
zgc:77026; eukaryotic translation initiation factor
5; K03262 translation initiation factor 5
Length=429
Score = 144 bits (363), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 64/130 (49%), Positives = 90/130 (69%), Gaps = 2/130 (1%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
+N+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTKFFGCELG
Sbjct 3 INVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKFFGCELG 62
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITV--KKGVLFCKC 129
A +F+ ++ ++NG+H L +LD FI+ +VLC C+ PE D+ + KK + C
Sbjct 63 AQTQFDSKNDRYIVNGSHEANKLQDMLDGFIRKFVLCPECDNPETDLHINPKKQTIGNSC 122
Query 130 NACGYSGTLD 139
ACGY G LD
Sbjct 123 KACGYRGMLD 132
> xla:380003 eif5, MGC52788; eukaryotic translation initiation
factor 5; K03262 translation initiation factor 5
Length=434
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 64/130 (49%), Positives = 89/130 (68%), Gaps = 2/130 (1%)
Query 13 NIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGA 72
N+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELGA
Sbjct 4 NVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELGA 63
Query 73 MVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITV--KKGVLFCKCN 130
+F+ ++ ++NG+H L +LD FI+ +VLC C+ PE D+ V KK + C
Sbjct 64 QTQFDIKNDRFIVNGSHEANKLQDMLDGFIKKFVLCPECDNPETDLHVNPKKQTIGNSCK 123
Query 131 ACGYSGTLDN 140
ACGY G LD
Sbjct 124 ACGYRGMLDT 133
> ath:AT1G36730 eukaryotic translation initiation factor 5, putative
/ eIF-5, putative; K03262 translation initiation factor
5
Length=439
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 66/124 (53%), Positives = 87/124 (70%), Gaps = 1/124 (0%)
Query 17 DKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 76
+++D YRYKMP++++KIEGRGNGI+TNV NM EIA+AL RP Y TK+FGCELGA KF
Sbjct 10 NRDDAFYRYKMPRMMTKIEGRGNGIKTNVVNMVEIAKALGRPAAYTTKYFGCELGAQSKF 69
Query 77 EENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKG-VLFCKCNACGYS 135
+E +L+NGAH L +L+ FI+ YV C GC PE +I + K +L KC ACG+
Sbjct 70 DEKNGTSLVNGAHDTSKLAGLLENFIKKYVQCYGCGNPETEILITKTQMLQLKCAACGFL 129
Query 136 GTLD 139
+D
Sbjct 130 SDVD 133
> ath:AT1G77840 eukaryotic translation initiation factor 5, putative
/ eIF-5, putative; K03262 translation initiation factor
5
Length=437
Score = 135 bits (339), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 62/124 (50%), Positives = 85/124 (68%), Gaps = 1/124 (0%)
Query 17 DKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 76
++ D YRYKMPK+++K EG+GNGI+TN+ N EIA+AL RPP Y TK+FGCELGA KF
Sbjct 10 NRNDAFYRYKMPKMVTKTEGKGNGIKTNIINNVEIAKALARPPSYTTKYFGCELGAQSKF 69
Query 77 EENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLF-CKCNACGYS 135
+E +L+NGAH+ L +L+ FI+ +V C GC PE +I + K + KC ACG+
Sbjct 70 DEKTGTSLVNGAHNTSKLAGLLENFIKKFVQCYGCGNPETEIIITKTQMVNLKCAACGFI 129
Query 136 GTLD 139
+D
Sbjct 130 SEVD 133
> sce:YPR041W TIF5, SUI5; Tif5p; K03262 translation initiation
factor 5
Length=405
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 80/129 (62%), Gaps = 1/129 (0%)
Query 12 VNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 71
+NI RD DP YRYKMP + +K+EGRGNGI+T V N+ +I+ AL RP Y K+FG ELG
Sbjct 3 INICRDNHDPFYRYKMPPIQAKVEGRGNGIKTAVLNVADISHALNRPAPYIVKYFGFELG 62
Query 72 AMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITV-KKGVLFCKCN 130
A ++++ L+NG H L +LD FI +VLCG C+ PE +I + K L C
Sbjct 63 AQTSISVDKDRYLVNGVHEPAKLQDVLDGFINKFVLCGSCKNPETEIIITKDNDLVRDCK 122
Query 131 ACGYSGTLD 139
ACG +D
Sbjct 123 ACGKRTPMD 131
> cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiation
factor) family member (iftb-1); K03238 translation initiation
factor 2 subunit 2
Length=250
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 51/111 (45%), Gaps = 6/111 (5%)
Query 23 YRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEK 82
+ K+P++ R +T N EI R +KR + +F ELG +
Sbjct 114 FAIKLPEV-----ARAGSKKTAFSNFLEICRLMKRQDKHVLQFLLAELGTTGSID-GSNC 167
Query 83 ALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCKCNACG 133
++ G ++ ++L K+I+ YV+C C+ PE +T + F +C CG
Sbjct 168 LIVKGRWQQKQFESVLRKYIKEYVMCHTCKSPETQLTKDTRLFFLQCTNCG 218
> ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238
translation initiation factor 2 subunit 2
Length=268
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query 40 GIRTNVY-NMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAIL 98
G + V+ N ++ + + R P + ++ ELG + +++ ++ G + ++ IL
Sbjct 154 GTKKTVFVNFMDLCKTMHRQPDHVMQYLLAELGTSGSLD-GQQRLVVKGRFAPKNFEGIL 212
Query 99 DKFIQMYVLCGGCELPEIDITVKKGVLFCKCNACG 133
++I YV+C GC+ P+ ++ + + F +C CG
Sbjct 213 RRYITDYVICLGCKSPDTILSKENRLFFLRCEKCG 247
> ath:AT3G07920 translation initiation factor; K03238 translation
initiation factor 2 subunit 2
Length=164
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 54/106 (50%), Gaps = 2/106 (1%)
Query 28 PKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALING 87
P++L + +G +T N + + + R P + F ELG K + ++++ ++ G
Sbjct 46 PQVLREETAKGTK-KTVFVNFMDYCKTMHRNPDHVMAFLLAELGTRGKLD-DQQRLVVRG 103
Query 88 AHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLFCKCNACG 133
+++D ++L +I YV+C C+ E ++ + + F +C CG
Sbjct 104 RFTQRDFESLLRGYILDYVMCIPCKSTEAILSKENRLFFLRCGKCG 149
> xla:779195 hypothetical protein MGC130708
Length=332
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 215 KTSFVNFTDICKLLHRQPKHLLAFLLAELGTSGSIDGNNQ-LVIKGRFQQKQIENVLRRY 273
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 274 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 304
> hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC8508;
eukaryotic translation initiation factor 2, subunit 2 beta,
38kDa; K03238 translation initiation factor 2 subunit 2
Length=333
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 216 KTSFVNFTDICKLLHRQPKHLLAFLLAELGTSGSIDGN-NQLVIKGRFQQKQIENVLRRY 274
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 275 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 305
> dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryotic
translation initiation factor 2, subunit 2 beta; K03238
translation initiation factor 2 subunit 2
Length=327
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 210 KTSFVNFTDICKLLHRQPKHLLVFLLAELGTSGSIDGNNQ-LVIKGRFQQKQIENVLRRY 268
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 269 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 299
> mmu:100040220 Gm9892; predicted gene 9892
Length=331
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 214 KTSFVNFTDICKLLHRQPKHLLAFLLAELGTSGSIDGN-NQLVIKGRFQQKQIENVLRRY 272
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 273 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 303
> mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381,
AA986487, AW822225, D2Ertd303e, EIF2, EIF2B, MGC130606; eukaryotic
translation initiation factor 2, subunit 2 (beta); K03238
translation initiation factor 2 subunit 2
Length=331
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 214 KTSFVNFTDICKLLHRQPKHLLAFLLAELGTSGSIDGN-NQLVIKGRFQQKQIENVLRRY 272
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 273 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 303
> xla:779197 eif2s2, MGC130959; eukaryotic translation initiation
factor 2, subunit 2 beta, 38kDa; K03238 translation initiation
factor 2 subunit 2
Length=335
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 218 KTSFVNFTDICKLLHRQPKHLLAFLLAELGTCGSIDGN-NQLVIKGRFQQKQIENVLRRY 276
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 277 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 307
> xla:780741 eukaryote initiation factor 2 beta
Length=335
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 42/91 (46%), Gaps = 1/91 (1%)
Query 42 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKF 101
+T+ N +I + L R P + F ELG + N + +I G ++ + +L ++
Sbjct 218 KTSFVNFTDICKLLHRQPKHLLAFLLAELGTCGSIDGN-NQLVIKGRFQQKQIENVLRRY 276
Query 102 IQMYVLCGGCELPEIDITVKKGVLFCKCNAC 132
I+ YV C C P+ + + F +C C
Sbjct 277 IKEYVTCHTCRSPDTILQKDTRLYFLQCETC 307
> sce:YPL237W SUI3; Beta subunit of the translation initiation
factor eIF2, involved in the identification of the start codon;
proposed to be involved in mRNA binding; K03238 translation
initiation factor 2 subunit 2
Length=285
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/126 (22%), Positives = 58/126 (46%), Gaps = 9/126 (7%)
Query 17 DKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 76
D+ P +R P L +G +T N+ +IA L R P + ++ ELG
Sbjct 152 DRSGPKFRIPPPVCLR------DGKKTIFSNIQDIAEKLHRSPEHLIQYLFAELGTSGSV 205
Query 77 EENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKG--VLFCKCNACGY 134
+ +++ +I G + + +L ++I YV C C+ ++ ++ + F C +CG
Sbjct 206 -DGQKRLVIKGKFQSKQMENVLRRYILEYVTCKTCKSINTELKREQSNRLFFMVCKSCGS 264
Query 135 SGTLDN 140
+ ++ +
Sbjct 265 TRSVSS 270
> ath:AT5G01940 eukaryotic translation initiation factor 2B family
protein / eIF-2B family protein; K03238 translation initiation
factor 2 subunit 2
Length=231
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/119 (22%), Positives = 54/119 (45%), Gaps = 7/119 (5%)
Query 8 KMQYVNIPRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFG 67
+++ ++ E P P+LL++ G T N ++ R + R P + KF
Sbjct 85 RLREEDVEVSTERPRTVMMPPQLLAE------GTITVCLNFADLCRTMHRKPDHVMKFLL 138
Query 68 CELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKKGVLF 126
++ V + +++ I G S +D A+ K+I +V+C C+ P+ + + LF
Sbjct 139 AQMETKVSLNK-QQRLEIKGLVSSKDFQAVFRKYIDAFVICVCCKSPDTALAEEGNGLF 196
> pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,
putative; K03238 translation initiation factor 2 subunit
2
Length=222
Score = 33.1 bits (74), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 38/89 (42%), Gaps = 3/89 (3%)
Query 47 NMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYV 106
N +I + R + F ELG E + ++ G + + + A+L K+I YV
Sbjct 108 NFKDICTIMNRNEEHVFHFVLAELGTEGSIA-GEGQLVLKGKYGPKHIEALLRKYITEYV 166
Query 107 LCGGCELPEIDI--TVKKGVLFCKCNACG 133
C C+ P + + + CNACG
Sbjct 167 TCQMCKSPNTTMEKDSRTRLFHQHCNACG 195
> tgo:TGME49_035540 translation initiation factor 2 beta, putative
; K03238 translation initiation factor 2 subunit 2
Length=231
Score = 31.6 bits (70), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 29/133 (21%), Positives = 54/133 (40%), Gaps = 14/133 (10%)
Query 9 MQYVNIPRDKEDPN------YRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYP 62
+Q + +K +P+ Y K P+++ R + N +I + R P +
Sbjct 79 LQRIQTLIEKHNPDLSGAKRYTIKPPQVV-----RVGSKKVAWINFKDICNIMNRQPDHV 133
Query 63 TKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYVLCGGCELPEIDITVKK 122
+F ELG + + ++ G + + + A+L K+I YV C C+ P +
Sbjct 134 HQFVLAELGTEGSIA-GDGQLVLKGKYGPKHIEALLRKYITEYVTCQMCKSPNTTMQRDS 192
Query 123 GVLFCK--CNACG 133
+ C ACG
Sbjct 193 RTRLWQQSCVACG 205
> tpv:TP02_0678 eukaryotic translation initiation factor 2 subunit
beta; K03238 translation initiation factor 2 subunit 2
Length=241
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 45/95 (47%), Gaps = 3/95 (3%)
Query 47 NMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYV 106
N +I + R + +F ELG + + ++ G + +++ ++L K+I YV
Sbjct 129 NFRDICSIMGRSMDHVHQFVLAELGTEGSIA-GDGQLVLKGKYGPKNIESLLRKYITEYV 187
Query 107 LCGGCELPEIDITVK-KGVLFCK-CNACGYSGTLD 139
C C+ P + + LF + C ACG + +++
Sbjct 188 TCSMCKSPNTTMERDCRARLFTQHCEACGANRSVN 222
> cpv:cgd8_4430 translation initiation factor if-2 betam beta
subunit ZnR ; K03238 translation initiation factor 2 subunit
2
Length=214
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/96 (22%), Positives = 43/96 (44%), Gaps = 3/96 (3%)
Query 47 NMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYV 106
N EI ++R + +F ELG + + ++ G + + + +L K+I YV
Sbjct 101 NFQEICNIMQRNADHVFQFVLSELGTEGSIA-GDGQLVLKGKYGPKHIEVLLRKYIMEYV 159
Query 107 LCGGCELP--EIDITVKKGVLFCKCNACGYSGTLDN 140
C C+ P ++ + + C ACG + ++ N
Sbjct 160 ACSMCKSPNTRLERDNRTRLYTIVCAACGANRSVQN 195
> hsa:84450 ZNF512, FLJ51176, KIAA1805, MGC111046; zinc finger
protein 512
Length=567
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 15 PRDKEDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMY 61
PR +ED +YR + P+ K+ GR +T + R K PP+Y
Sbjct 105 PRQEEDEDYR-EFPQKKHKLYGRKQRPKTQPNPKSQARRIRKEPPVY 150
> pfa:PF14_0315 conserved Plasmodium membrane protein, unknown
function
Length=5757
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 32/74 (43%), Gaps = 14/74 (18%)
Query 19 EDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEE 78
ED YK+P ++ + R N I TN+Y+ K F +L F++
Sbjct 905 EDRRIIYKLPVIILNLPSRKNKIYTNIYSC--------------EKHFNTKLYCFFIFKQ 950
Query 79 NEEKALINGAHSEQ 92
+++G H+E+
Sbjct 951 RNNNEIVDGVHNEK 964
> dre:100333914 C-C motif chemokine 21-like
Length=156
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 10/19 (52%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 109 GGCELPEIDITVKKGVLFC 127
GGC +P I T+K+G +FC
Sbjct 53 GGCNIPAIVFTLKQGRMFC 71
> mmu:229503 Rrnad1, MGC38548; ribosomal RNA adenine dimethylase
domain containing 1
Length=475
Score = 28.1 bits (61), Expect = 8.3, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 63 TKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDK 100
++F LG MVK E ++ + H +Q+L+ LDK
Sbjct 168 SRFMSLGLGLMVKSLEGNQRLVKRAQHLDQELLKALDK 205
> bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation
factor 2, beta; K03238 translation initiation factor 2 subunit
2
Length=238
Score = 28.1 bits (61), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 21/95 (22%), Positives = 44/95 (46%), Gaps = 3/95 (3%)
Query 47 NMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEQDLVAILDKFIQMYV 106
N E+ + R + +F ELG + + ++ G + +++ ++L K+I YV
Sbjct 126 NFKELCNIMGRTMDHVHQFVLAELGTEGSIA-GDGQLVLKGKYGPKNIESLLRKYITEYV 184
Query 107 LCGGCELPEIDITV-KKGVLFCK-CNACGYSGTLD 139
C C+ + + LF + C ACG + +++
Sbjct 185 TCSMCKSANTTMERDSRARLFTQHCEACGANRSVN 219
Lambda K H
0.321 0.140 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40