bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0777_orf2
Length=235
Score E
Sequences producing significant alignments: (Bits) Value
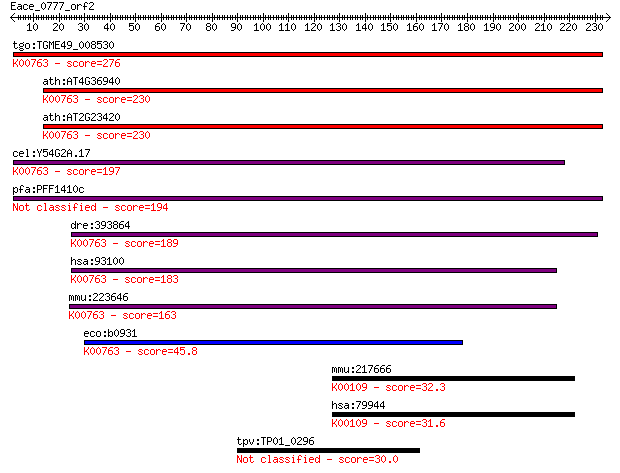
tgo:TGME49_008530 nicotinate phosphoribosyltransferase, putati... 276 3e-74
ath:AT4G36940 NAPRT1; NAPRT1 (NICOTINATE PHOSPHORIBOSYLTRANSFE... 230 3e-60
ath:AT2G23420 NAPRT2; NAPRT2 (NICOTINATE PHOSPHORIBOSYLTRANSFE... 230 3e-60
cel:Y54G2A.17 hypothetical protein; K00763 nicotinate phosphor... 197 2e-50
pfa:PFF1410c conserved protein, unknown function 194 2e-49
dre:393864 naprt1, MGC77870, zgc:77870; nicotinate phosphoribo... 189 9e-48
hsa:93100 NAPRT1, PP3856; nicotinate phosphoribosyltransferase... 183 5e-46
mmu:223646 Naprt1, 9130210N20Rik; nicotinate phosphoribosyltra... 163 4e-40
eco:b0931 pncB, ECK0922, JW0914; nicotinate phosphoribosyltran... 45.8 1e-04
mmu:217666 L2hgdh, BC016226, MGC28775; L-2-hydroxyglutarate de... 32.3 1.5
hsa:79944 L2HGDH, C14orf160, DURANIN, FLJ12618; L-2-hydroxyglu... 31.6 2.6
tpv:TP01_0296 hypothetical protein 30.0 7.9
> tgo:TGME49_008530 nicotinate phosphoribosyltransferase, putative
(EC:2.4.2.11); K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=589
Score = 276 bits (707), Expect = 3e-74, Method: Compositional matrix adjust.
Identities = 130/232 (56%), Positives = 166/232 (71%), Gaps = 1/232 (0%)
Query 2 DFPQAVMSARDLLFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIP 61
D + V+ R + +WP N M GELAAF A+A TFP+ FLALVDTY+TL SG+P
Sbjct 284 DIIERVLKYRQKVIDLWPSENLDSMMNMGELAAFTAFAQTFPNAFLALVDTYDTLCSGVP 343
Query 62 NFLAVALAMFKLGAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP-FGGLSIVLSNDL 120
N L V+ A+ + G P+G+R+DSGDLAYLSRE R++F + AF P G L I SNDL
Sbjct 344 NALVVSAALLECGYHPRGIRLDSGDLAYLSREVRKLFHEAAAAFEMPDLGRLKIAASNDL 403
Query 121 NEATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLP 180
NE I+++ DEGHE D+F IGTN+VTCQAQPALG+VYKLVEL+G CMK+S+ EK SLP
Sbjct 404 NEVVISSVRDEGHEIDIFAIGTNLVTCQAQPALGMVYKLVELDGAACMKVSQVFEKASLP 463
Query 181 TAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDLYADKRRCFFSPRR 232
K AYRL+ K+G PAVDL+Q A P PV G+++FC+ LY D++RCF P +
Sbjct 464 CKKEAYRLFTKDGAPAVDLLQEAKDPPPVEGKRIFCRHLYDDRKRCFLVPSK 515
> ath:AT4G36940 NAPRT1; NAPRT1 (NICOTINATE PHOSPHORIBOSYLTRANSFERASE
1); nicotinate phosphoribosyltransferase (EC:2.4.2.11);
K00763 nicotinate phosphoribosyltransferase [EC:2.4.2.11]
Length=559
Score = 230 bits (587), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 115/220 (52%), Positives = 150/220 (68%), Gaps = 7/220 (3%)
Query 14 LFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKL 73
L G++ E N + ELAAF +YA+ FP++FLALVDTY+ + SGIPNF AVALA+ +L
Sbjct 270 LQGIFSETN------QSELAAFISYALAFPNSFLALVDTYDVMKSGIPNFCAVALALNEL 323
Query 74 GAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP-FGGLSIVLSNDLNEATITALNDEG 132
G K G+R+DSGDLAYLS E R+ F E P FG + + SNDLNE T+ ALN +G
Sbjct 324 GYKAVGIRLDSGDLAYLSTEVRKFFCAIERDLKVPDFGKMIVTASNDLNEETVDALNKQG 383
Query 133 HEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKE 192
HE D FGIGTN+VTC AQ ALG V+KLVE+ +P +KLSEDV K S+P K YRL+ KE
Sbjct 384 HEVDAFGIGTNLVTCYAQAALGCVFKLVEINNQPRIKLSEDVTKVSIPCKKRTYRLFGKE 443
Query 193 GVPAVDLIQSASMPRPVCGEKLFCKDLYADKRRCFFSPRR 232
G P VD++ + P P GE+L C+ + + +R + P+R
Sbjct 444 GYPLVDIMTGENEPPPKVGERLLCRHPFNESKRAYVVPQR 483
> ath:AT2G23420 NAPRT2; NAPRT2 (NICOTINATE PHOSPHORIBOSYLTRANSFERASE
2); nicotinate phosphoribosyltransferase (EC:2.4.2.11);
K00763 nicotinate phosphoribosyltransferase [EC:2.4.2.11]
Length=557
Score = 230 bits (586), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 116/220 (52%), Positives = 147/220 (66%), Gaps = 7/220 (3%)
Query 14 LFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKL 73
L G++ E N + ELAAF +YA+ FP FLALVDTY+ + SGIPNF AVALA+
Sbjct 268 LSGIFSETN------QSELAAFTSYALAFPKTFLALVDTYDVMKSGIPNFCAVALALNDF 321
Query 74 GAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP-FGGLSIVLSNDLNEATITALNDEG 132
G K G+R+DSGDLAYLSREAR F E P FG + + SNDLNE TI ALN +G
Sbjct 322 GYKALGIRLDSGDLAYLSREARNFFCTVERELKVPGFGKMVVTASNDLNEETIDALNKQG 381
Query 133 HEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKE 192
HE D FGIGT +VTC +Q ALG V+KLVE+ +P +KLSEDV K S+P K +YRLY KE
Sbjct 382 HEVDAFGIGTYLVTCYSQAALGCVFKLVEINNQPRIKLSEDVTKVSIPCKKRSYRLYGKE 441
Query 193 GVPAVDLIQSASMPRPVCGEKLFCKDLYADKRRCFFSPRR 232
G P VD++ + P P GE+L C+ + + +R + P+R
Sbjct 442 GYPLVDIMTGENEPPPKVGERLLCRHPFNESKRAYVVPQR 481
> cel:Y54G2A.17 hypothetical protein; K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=563
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 100/218 (45%), Positives = 136/218 (62%), Gaps = 2/218 (0%)
Query 2 DFPQAVMSARDLLFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIP 61
D Q M R L + +GEL+AF AYA+ FPD FLAL+DTY+ + SG+
Sbjct 255 DLFQISMEKRAWLLDQFSWKAALSEVSDGELSAFVAYAIAFPDTFLALIDTYDVIRSGVV 314
Query 62 NFLAVALAMFKLGAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFP--FGGLSIVLSND 119
NF+AV+LA+ LG + G RIDSGDL+YLS+E R F + G F +SIV SND
Sbjct 315 NFVAVSLALHDLGYRSMGCRIDSGDLSYLSKELRECFVKVSTLKGEYKFFEKMSIVASND 374
Query 120 LNEATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSL 179
+NE TI +LND+ HE + FG+GT++VTCQ QPALG VYKLV +P +KLS+DV K ++
Sbjct 375 INEETIMSLNDQQHEINAFGVGTHLVTCQKQPALGCVYKLVAQSAQPKIKLSQDVTKITI 434
Query 180 PTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCK 217
P K YR++ K G +DL+ P P +++ C+
Sbjct 435 PGKKKCYRIFGKNGYAILDLMMLEDEPEPQPNQQILCR 472
> pfa:PFF1410c conserved protein, unknown function
Length=665
Score = 194 bits (494), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 85/231 (36%), Positives = 142/231 (61%), Gaps = 4/231 (1%)
Query 2 DFPQAVMSARDLLFGVWPEANFHQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIP 61
DF + ++++ ++ + E EL AF+A+A P NF+ L+DTY+TL SGI
Sbjct 367 DFVSIINKNKNIVHKLYD----CKYTNESELIAFSAFAQINPKNFICLIDTYDTLKSGIY 422
Query 62 NFLAVALAMFKLGAKPKGVRIDSGDLAYLSREARRMFKQCEEAFGFPFGGLSIVLSNDLN 121
NFL VAL+++++ KP G+RIDSGDL L+ E + +F + PF L I +SND+N
Sbjct 423 NFLIVALSLYEINYKPIGIRIDSGDLRSLTNECKNIFNDISQKLNVPFNQLKICVSNDIN 482
Query 122 EATITALNDEGHEADVFGIGTNVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPT 181
+ I LN+E + D+F IGTN++TCQ+QP+LG+VYKLV++ PC K++ + +K++LP
Sbjct 483 QKIINYLNEEQNHIDIFAIGTNLITCQSQPSLGLVYKLVQINNAPCFKMTNENKKSNLPY 542
Query 182 AKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDLYADKRRCFFSPRR 232
K YR+Y + +D IQ + P+ ++ +++ + ++ P++
Sbjct 543 QKNVYRIYTDQQFATLDYIQLYNEQPPILNQQTISINIHDEWKQTIIIPKK 593
> dre:393864 naprt1, MGC77870, zgc:77870; nicotinate phosphoribosyltransferase
domain containing 1 (EC:2.4.2.11); K00763 nicotinate
phosphoribosyltransferase [EC:2.4.2.11]
Length=523
Score = 189 bits (479), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 95/208 (45%), Positives = 139/208 (66%), Gaps = 2/208 (0%)
Query 25 QMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKLGAKPKGVRIDS 84
+ +EGELAAF +YA+ +P NFL ++D+Y+ SG+ F AVALA+F+L +P GVR+DS
Sbjct 237 NLIREGELAAFLSYAIAYPQNFLPVIDSYSVRCSGLLCFCAVALALFELQYRPLGVRLDS 296
Query 85 GDLAYLSREARRMFKQCEEAFGFP-FGGLSIVLSNDLNEATITALNDEGHEADVFGIGTN 143
GDL S + RR+F++C + F P F L IV +N ++E +++ LN + +E DV G+GT+
Sbjct 297 GDLCRQSLDVRRVFRECSKHFSVPQFESLIIVGTNSISEQSLSELNKKDNEIDVVGVGTH 356
Query 144 VVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKEGVPAVDLIQSA 203
+VTC QP+LG VYKLVE+ G+P MKLSED EK++LP KA YRL + +G P +D++
Sbjct 357 LVTCTRQPSLGCVYKLVEVRGRPRMKLSEDPEKSTLPGRKAVYRLLDADGHPQMDVLCLC 416
Query 204 SMPRPVCGEKLFCKDLYADKR-RCFFSP 230
P G +L C L + +C +P
Sbjct 417 DEAAPAAGVELQCFSLLGNSTLQCSITP 444
> hsa:93100 NAPRT1, PP3856; nicotinate phosphoribosyltransferase
domain containing 1 (EC:2.4.2.11); K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=538
Score = 183 bits (464), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 94/191 (49%), Positives = 129/191 (67%), Gaps = 1/191 (0%)
Query 25 QMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKLGAKPKGVRIDS 84
Q GE AAF AYA+ FP F L+DTY+ SG+PNFLAVALA+ +LG + GVR+DS
Sbjct 262 QEPHPGERAAFVAYALAFPRAFQGLLDTYSVWRSGLPNFLAVALALGELGYRAVGVRLDS 321
Query 85 GDLAYLSREARRMFKQCEEAFGFPF-GGLSIVLSNDLNEATITALNDEGHEADVFGIGTN 143
GDL ++E R++F+ F P+ + IV+SN+++E + L EG E +V GIGT+
Sbjct 322 GDLLQQAQEIRKVFRAAAAQFQVPWLESVLIVVSNNIDEEALARLAQEGSEVNVIGIGTS 381
Query 144 VVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKEGVPAVDLIQSA 203
VVTC QP+LG VYKLV + G+P MKL+ED EK +LP +KAA+RL +G P +D++Q A
Sbjct 382 VVTCPQQPSLGGVYKLVAVGGQPRMKLTEDPEKQTLPGSKAAFRLLGSDGSPLMDMLQLA 441
Query 204 SMPRPVCGEKL 214
P P G++L
Sbjct 442 EEPVPQAGQEL 452
> mmu:223646 Naprt1, 9130210N20Rik; nicotinate phosphoribosyltransferase
domain containing 1 (EC:2.4.2.11); K00763 nicotinate
phosphoribosyltransferase [EC:2.4.2.11]
Length=538
Score = 163 bits (413), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 88/192 (45%), Positives = 126/192 (65%), Gaps = 1/192 (0%)
Query 24 HQMAKEGELAAFAAYAMTFPDNFLALVDTYNTLSSGIPNFLAVALAMFKLGAKPKGVRID 83
Q GE AAF AYA+ FP F L+D+Y+ SG+PNFLAVALA+ +LG + GVR+D
Sbjct 261 EQEPHPGERAAFVAYALAFPRAFQGLLDSYSVRRSGLPNFLAVALALGELGYRAVGVRLD 320
Query 84 SGDLAYLSREARRMFKQCEEAFGFPF-GGLSIVLSNDLNEATITALNDEGHEADVFGIGT 142
SGDL ++E R +F+ F P+ + I +SN+++E+ + L +G E +V GIGT
Sbjct 321 SGDLLQQAKEIRGIFRTAGAQFQMPWLESVPIAVSNNIDESELMRLAQKGSEVNVIGIGT 380
Query 143 NVVTCQAQPALGVVYKLVELEGKPCMKLSEDVEKTSLPTAKAAYRLYNKEGVPAVDLIQS 202
+VVTC QP++G VYKLV + G+P +KL+ED EK +LP +KAA+R +G +DL+Q
Sbjct 381 SVVTCPKQPSMGCVYKLVSVGGQPRIKLTEDPEKQTLPGSKAAFRFLGPDGSLLLDLLQL 440
Query 203 ASMPRPVCGEKL 214
A P P G++L
Sbjct 441 AEEPPPKAGQEL 452
> eco:b0931 pncB, ECK0922, JW0914; nicotinate phosphoribosyltransferase
(EC:2.4.2.11); K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=400
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 48/154 (31%), Positives = 72/154 (46%), Gaps = 19/154 (12%)
Query 30 GELAAFAAYAMTFPDNF-LALVDTYNTLSSGIPNFLAVALAMFKLGAKPKGVRIDSGDLA 88
+ AA AA+ +PD +AL D T+ + + +F + ++ +G+R DSGD
Sbjct 236 SQRAALAAWLEEYPDQLGIALTDCI-TMDAFLRDF------GVEFASRYQGLRHDSGDPV 288
Query 89 YLSREARRMFKQCEEAFGFPFGGLSIVLSN--DLNEATITALNDEGHEADVFGIGTNVVT 146
+A + E G ++V S+ DL +A + FGIGT + T
Sbjct 289 EWGEKAIAHY----EKLGIDPQSKTLVFSDNLDLRKAVELYRHFSSRVQLSFGIGTRL-T 343
Query 147 C---QAQPALGVVYKLVELEGKPCMKLSEDVEKT 177
C Q +P L +V KLVE GKP KLS+ KT
Sbjct 344 CDIPQVKP-LNIVIKLVECNGKPVAKLSDSPGKT 376
> mmu:217666 L2hgdh, BC016226, MGC28775; L-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.2); K00109 2-hydroxyglutarate dehydrogenase
[EC:1.1.99.2]
Length=464
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 49/103 (47%), Gaps = 12/103 (11%)
Query 127 ALNDEGHEADVF--GIGTNVVTCQAQPAL---GVVYKLVELEGKP---CMKLSEDVEKTS 178
AL+ GH + V GI + +A+ + ++Y+ L+G P C KL VE+
Sbjct 87 ALHQTGHNSGVIHSGIYYKPESLKAKLCVEGAALIYEYCNLKGIPYRQCGKLIVAVEQEE 146
Query 179 LPTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDLYA 221
+P +A Y + GV + LIQ + + ++ +C+ L A
Sbjct 147 IPRLQALYERGLQNGVEGLRLIQQEDIKK----KEPYCRGLMA 185
> hsa:79944 L2HGDH, C14orf160, DURANIN, FLJ12618; L-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.2); K00109 2-hydroxyglutarate
dehydrogenase [EC:1.1.99.2]
Length=463
Score = 31.6 bits (70), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 46/105 (43%), Gaps = 16/105 (15%)
Query 127 ALNDEGHEADVFGIGT-------NVVTCQAQPALGVVYKLVELEG---KPCMKLSEDVEK 176
A++ GH + V G C AL +Y+ + +G K C KL VE+
Sbjct 86 AVHQTGHNSGVIHSGIYYKPESLKAKLCVQGAAL--LYEYCQQKGISYKQCGKLIVAVEQ 143
Query 177 TSLPTAKAAYRLYNKEGVPAVDLIQSASMPRPVCGEKLFCKDLYA 221
+P +A Y + GVP + LIQ + + ++ +C+ L A
Sbjct 144 EEIPRLQALYEKGLQNGVPGLRLIQQEDIKK----KEPYCRGLMA 184
> tpv:TP01_0296 hypothetical protein
Length=402
Score = 30.0 bits (66), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 16/71 (22%), Positives = 35/71 (49%), Gaps = 0/71 (0%)
Query 90 LSREARRMFKQCEEAFGFPFGGLSIVLSNDLNEATITALNDEGHEADVFGIGTNVVTCQA 149
++R+ R+ K+ E+A + +++S + E T LN E A++ + +C
Sbjct 330 VNRDQYRLAKENEDAIHRNYNDQLVLMSEHITELNNTILNSEYKIAELTQTKVSCPSCGF 389
Query 150 QPALGVVYKLV 160
LG++Y ++
Sbjct 390 NNTLGILYTVL 400
Lambda K H
0.321 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8081806824
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40