bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0794_orf1
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
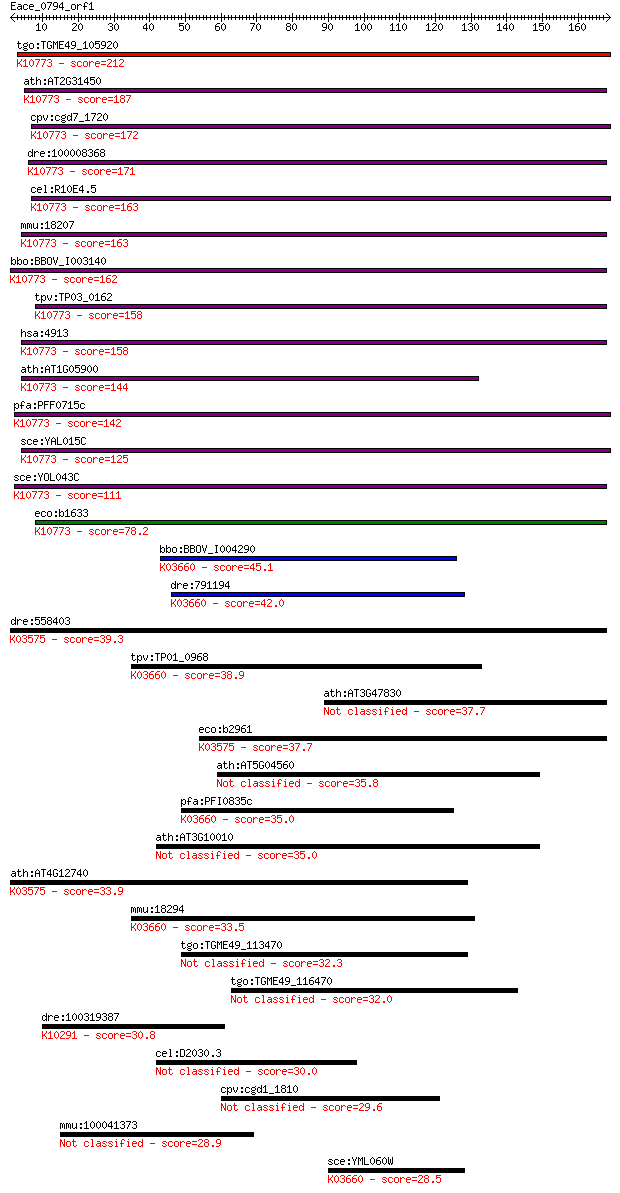
tgo:TGME49_105920 endonuclease III-like protein 1, putative (E... 212 5e-55
ath:AT2G31450 endonuclease-related; K10773 endonuclease III [E... 187 1e-47
cpv:cgd7_1720 endonuclease III ; K10773 endonuclease III [EC:4... 172 7e-43
dre:100008368 RUN domain containing 3A; K10773 endonuclease II... 171 1e-42
cel:R10E4.5 nth-1; NTH (eNdonuclease THree like) homolog famil... 163 3e-40
mmu:18207 Nthl1, Nth1, Octs3; nth (endonuclease III)-like 1 (E... 163 3e-40
bbo:BBOV_I003140 19.m02342; base excision DNA repair protein, ... 162 7e-40
tpv:TP03_0162 endonuclease III (EC:4.2.99.18); K10773 endonucl... 158 7e-39
hsa:4913 NTHL1, NTH1, OCTS3; nth endonuclease III-like 1 (E. c... 158 1e-38
ath:AT1G05900 endonuclease-related; K10773 endonuclease III [E... 144 2e-34
pfa:PFF0715c endonuclease III homologue, putative (EC:4.2.99.1... 142 5e-34
sce:YAL015C NTG1, FUN33, SCR1; DNA N-glycosylase and apurinic/... 125 7e-29
sce:YOL043C NTG2, SCR2; DNA N-glycosylase and apurinic/apyrimi... 111 1e-24
eco:b1633 nth, ECK1629, JW1625; DNA glycosylase and apyrimidin... 78.2 1e-14
bbo:BBOV_I004290 19.m02329; 8-oxoguanine DNA glycosylase; K036... 45.1 1e-04
dre:791194 ogg1, zgc:158858; 8-oxoguanine DNA glycosylase (EC:... 42.0 0.001
dre:558403 mutyh, si:ch211-241d21.4; mutY homolog (E. coli); K... 39.3 0.007
tpv:TP01_0968 8-oxoguanine DNA-glycosylase; K03660 N-glycosyla... 38.9 0.008
ath:AT3G47830 HhH-GPD base excision DNA repair protein-related 37.7 0.018
eco:b2961 mutY, ECK2956, JW2928, micA, mutB; adenine DNA glyco... 37.7 0.018
ath:AT5G04560 DME; DME (DEMETER); DNA N-glycosylase/ DNA-(apur... 35.8 0.070
pfa:PFI0835c N-glycosylase/DNA lyase, putative (EC:3.2.2.- 4.2... 35.0 0.12
ath:AT3G10010 DML2; DML2 (DEMETER-LIKE 2); 4 iron, 4 sulfur cl... 35.0 0.12
ath:AT4G12740 adenine-DNA glycosylase-related / MYH-related; K... 33.9 0.28
mmu:18294 Ogg1, Mmh; 8-oxoguanine DNA-glycosylase 1 (EC:4.2.99... 33.5 0.35
tgo:TGME49_113470 helix-hairpin-helix motif-containing protein... 32.3 0.77
tgo:TGME49_116470 hypothetical protein 32.0 1.1
dre:100319387 si:dkey-46a10.2; K10291 F-box protein 4 30.8
cel:D2030.3 hypothetical protein 30.0 3.5
cpv:cgd1_1810 hypothetical protein 29.6 5.5
mmu:100041373 Gm3297; predicted gene 3297 28.9
sce:YML060W OGG1; Mitochondrial glycosylase/lyase that specifi... 28.5 9.9
> tgo:TGME49_105920 endonuclease III-like protein 1, putative
(EC:4.2.99.18); K10773 endonuclease III [EC:4.2.99.18]
Length=523
Score = 212 bits (539), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 100/166 (60%), Positives = 129/166 (77%), Gaps = 0/166 (0%)
Query 3 EKERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHN 62
EK +RF+VLVAVMLSSQTKDEQT AC+ RLR+ +L+P ++ +L LL+GVGF+
Sbjct 318 EKAKRFSVLVAVMLSSQTKDEQTAACMQRLRDADVLSPEKMSRLSVAELSELLYGVGFYQ 377
Query 63 NKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVH 122
NK FLKEA +IL++K+ G++P T ++L+QL+GVG KMANI +HA WN GIAVDVHVH
Sbjct 378 NKARFLKEACQILLEKYGGDIPPTYEELVQLKGVGPKMANIAVHAGWNRVEGIAVDVHVH 437
Query 123 RISNRLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
RI+NRL WV+TKTP ET+ AL+ L R W ++NLL VGFGQQIC+
Sbjct 438 RITNRLNWVRTKTPIETQHALQKFLRRPLWGEINLLFVGFGQQICR 483
> ath:AT2G31450 endonuclease-related; K10773 endonuclease III
[EC:4.2.99.18]
Length=377
Score = 187 bits (476), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 85/170 (50%), Positives = 119/170 (70%), Gaps = 7/170 (4%)
Query 5 ERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNK 64
ERRFAVL+ +LSSQTKD+ +A ++RL +NGLL P A+ D ++ L++ VGF+ K
Sbjct 169 ERRFAVLLGALLSSQTKDQVNNAAIHRLHQNGLLTPEAVDKADESTIKELIYPVGFYTRK 228
Query 65 TTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRI 124
T++K+ A I + K+ G++P +LDDL+ L G+G KMA++++H AWN GI VD HVHRI
Sbjct 229 ATYMKKIARICLVKYDGDIPSSLDDLLSLPGIGPKMAHLILHIAWNDVQGICVDTHVHRI 288
Query 125 SNRLGWV-------KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
NRLGWV KT +P+ET +AL+ LP++ W +N LLVGFGQ IC
Sbjct 289 CNRLGWVSRPGTKQKTTSPEETRVALQQWLPKEEWVAINPLLVGFGQMIC 338
> cpv:cgd7_1720 endonuclease III ; K10773 endonuclease III [EC:4.2.99.18]
Length=189
Score = 172 bits (435), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 83/162 (51%), Positives = 111/162 (68%), Gaps = 1/162 (0%)
Query 7 RFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNKTT 66
RF VLVA LSSQTKDE T AC+NRL +NGL +P I + LR +L+GVGF+N K
Sbjct 1 RFHVLVAAFLSSQTKDEVTAACMNRLIDNGL-SPEFINNQSVDSLRDMLYGVGFYNTKAK 59
Query 67 FLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISN 126
LKE + I+I+ + G+VP+ + L+ L G+G KMAN+++ + GI+VD H+HRI N
Sbjct 60 NLKEISRIIIQNYSGKVPEKYEQLVMLPGIGPKMANLILQIGFGIVVGISVDTHMHRIFN 119
Query 127 RLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
R+GWVKTK P ET +E LPR +W D+N + VG+GQ ICK
Sbjct 120 RIGWVKTKNPIETSKEMEKMLPRIYWNDINKVFVGYGQTICK 161
> dre:100008368 RUN domain containing 3A; K10773 endonuclease
III [EC:4.2.99.18]
Length=430
Score = 171 bits (432), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 84/164 (51%), Positives = 114/164 (69%), Gaps = 3/164 (1%)
Query 6 RRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNKT 65
RR+ VL+++MLSSQTKD+ T + RLRE+GL + I D E L L++ VGF K
Sbjct 166 RRYQVLISLMLSSQTKDQVTAGAMQRLREHGL-SVDGILKMDDETLGKLIYPVGFWRTKV 224
Query 66 TFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRIS 125
++K+A ++ ++ G++P T++ L++L GVG KMA++ M AWN GI VD HVHRIS
Sbjct 225 KYIKQATALIQQEFGGDIPNTVEGLIRLPGVGPKMAHLAMDIAWNQVSGIGVDTHVHRIS 284
Query 126 NRLGWVK--TKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
NRLGW K TKTP+ET ALE+ LPR W ++N LLVGFGQQ+C
Sbjct 285 NRLGWTKKETKTPEETRRALEEWLPRDLWSEINWLLVGFGQQVC 328
> cel:R10E4.5 nth-1; NTH (eNdonuclease THree like) homolog family
member (nth-1); K10773 endonuclease III [EC:4.2.99.18]
Length=259
Score = 163 bits (412), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 83/162 (51%), Positives = 109/162 (67%), Gaps = 1/162 (0%)
Query 7 RFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNKTT 66
RF VLVA+MLSSQT+DE A + RL+++GL + P+ L +L VGF+ K
Sbjct 28 RFQVLVALMLSSQTRDEVNAAAMKRLKDHGLSIGKILEFKVPD-LETILCPVGFYKRKAV 86
Query 67 FLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISN 126
+L++ A+IL G++P +LD L L GVG KMAN+VM AW GIAVD HVHRISN
Sbjct 87 YLQKTAKILKDDFSGDIPDSLDGLCALPGVGPKMANLVMQIAWGECVGIAVDTHVHRISN 146
Query 127 RLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
RLGW+KT TP++T+ ALE LP+ W+ +N LLVGFGQ C+
Sbjct 147 RLGWIKTSTPEKTQKALEILLPKSEWQPINHLLVGFGQMQCQ 188
> mmu:18207 Nthl1, Nth1, Octs3; nth (endonuclease III)-like 1
(E.coli) (EC:4.2.99.18); K10773 endonuclease III [EC:4.2.99.18]
Length=300
Score = 163 bits (412), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 83/166 (50%), Positives = 113/166 (68%), Gaps = 3/166 (1%)
Query 4 KERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNN 63
K RR+ VL+++MLSSQTKD+ T + RLR GL +I D + L L++ VGF N
Sbjct 114 KVRRYQVLLSLMLSSQTKDQVTAGAMQRLRARGL-TVESILQTDDDTLGRLIYPVGFWRN 172
Query 64 KTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHR 123
K ++K+ IL ++++G++P ++ +L+ L GVG KMA++ M AW + GIAVD HVHR
Sbjct 173 KVKYIKQTTAILQQRYEGDIPASVAELVALPGVGPKMAHLAMAVAWGTISGIAVDTHVHR 232
Query 124 ISNRLGWVK--TKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
I+NRL W K TKTP+ET LE+ LPR W +VN LLVGFGQQIC
Sbjct 233 IANRLRWTKKMTKTPEETRKNLEEWLPRVLWSEVNGLLVGFGQQIC 278
> bbo:BBOV_I003140 19.m02342; base excision DNA repair protein,
HhH-GPD family domain containing protein; K10773 endonuclease
III [EC:4.2.99.18]
Length=205
Score = 162 bits (409), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 78/167 (46%), Positives = 114/167 (68%), Gaps = 1/167 (0%)
Query 1 KDEKERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGF 60
+ E+ ++ L+A MLSSQTKD T A ++ L++ GL P I+ ++L L+ VGF
Sbjct 11 QSERIYQYQTLIACMLSSQTKDAVTAAAMDALKQRGL-TPENISKMPEDELDSLISKVGF 69
Query 61 HNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVH 120
H K +K+A E+++ K G+VP ++DL+ L GVG KM N+V+ + +GIAVD+H
Sbjct 70 HKTKAKHIKQATEMILNKFGGKVPDNIEDLVTLPGVGPKMGNLVLQIGFKRINGIAVDLH 129
Query 121 VHRISNRLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
VHRI+NRL WVKTKTP+ET I L++ +P++ W +VN LLVGFGQ +C
Sbjct 130 VHRIANRLQWVKTKTPEETRIKLQELIPKRLWAEVNHLLVGFGQTVC 176
> tpv:TP03_0162 endonuclease III (EC:4.2.99.18); K10773 endonuclease
III [EC:4.2.99.18]
Length=418
Score = 158 bits (400), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 79/160 (49%), Positives = 105/160 (65%), Gaps = 1/160 (0%)
Query 8 FAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNKTTF 67
F LV MLSSQTKDE T + L++ GL I D E+L ++ VGFH K
Sbjct 233 FQTLVGCMLSSQTKDEITALTMKNLKKRGL-TLDNILKMDEEELDSIISKVGFHKTKAKN 291
Query 68 LKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISNR 127
+K+AA+IL ++ G+VP DL L G+G KMAN+++ A+N G+AVD+HVHRI+NR
Sbjct 292 IKKAAQILKDQYGGKVPSNKKDLESLPGIGPKMANLILQVAFNMVDGVAVDIHVHRITNR 351
Query 128 LGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
LGWVKTKTP+ET + L++ LP+ W +N LLVGFGQ C
Sbjct 352 LGWVKTKTPEETSLKLQELLPKDLWSKINPLLVGFGQTFC 391
> hsa:4913 NTHL1, NTH1, OCTS3; nth endonuclease III-like 1 (E.
coli) (EC:4.2.99.18); K10773 endonuclease III [EC:4.2.99.18]
Length=312
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 80/166 (48%), Positives = 112/166 (67%), Gaps = 3/166 (1%)
Query 4 KERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNN 63
K RR+ VL+++MLSSQTKD+ T + RLR GL +I D L L++ VGF +
Sbjct 126 KVRRYQVLLSLMLSSQTKDQVTAGAMQRLRARGLTV-DSILQTDDATLGKLIYPVGFWRS 184
Query 64 KTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHR 123
K ++K+ + IL + + G++P ++ +L+ L GVG KMA++ M AW + GIAVD HVHR
Sbjct 185 KVKYIKQTSAILQQHYGGDIPASVAELVALPGVGPKMAHLAMAVAWGTVSGIAVDTHVHR 244
Query 124 ISNRLGWVK--TKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
I+NRL W K TK+P+ET ALE+ LPR+ W ++N LLVGFGQQ C
Sbjct 245 IANRLRWTKKATKSPEETRAALEEWLPRELWHEINGLLVGFGQQTC 290
> ath:AT1G05900 endonuclease-related; K10773 endonuclease III
[EC:4.2.99.18]
Length=314
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 66/128 (51%), Positives = 91/128 (71%), Gaps = 0/128 (0%)
Query 4 KERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNN 63
KERRF VL+ +LSSQTK+ T A V RL +NGLL P AI D ++ L++ VGF+
Sbjct 177 KERRFYVLIGTLLSSQTKEHITGAAVERLHQNGLLTPEAIDKADESTIKELIYPVGFYTR 236
Query 64 KTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHR 123
K T +K+ A+I + ++ G++P+TL++L+ L GVG K+A++V+H AWN GI VD HVHR
Sbjct 237 KATNVKKVAKICLMEYDGDIPRTLEELLSLPGVGPKIAHLVLHVAWNDVQGICVDTHVHR 296
Query 124 ISNRLGWV 131
I NRLGWV
Sbjct 297 ICNRLGWV 304
> pfa:PFF0715c endonuclease III homologue, putative (EC:4.2.99.18);
K10773 endonuclease III [EC:4.2.99.18]
Length=437
Score = 142 bits (359), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 68/167 (40%), Positives = 110/167 (65%), Gaps = 1/167 (0%)
Query 2 DEKERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFH 61
D K RF L++ MLSS+TKDE T +++L+++GL + I E+L+ L++G+GF+
Sbjct 239 DMKVFRFQTLISCMLSSRTKDEVTAMVMDKLKKHGL-TVHNILNTTEEQLKKLIYGIGFY 297
Query 62 NNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHV 121
N K + + IL K+ ++P T ++L +L G+G K+A +++ A N GIAVD+HV
Sbjct 298 NVKAKQILQICHILKNKYNSDIPHTYEELKKLPGIGEKIAQLILQTALNKHEGIAVDIHV 357
Query 122 HRISNRLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
HRI+NRL WV +K +T++ L+ + ++ W ++N +LVGFGQ ICK
Sbjct 358 HRIANRLNWVNSKNELDTQMKLKSYVQKELWSEINHVLVGFGQVICK 404
> sce:YAL015C NTG1, FUN33, SCR1; DNA N-glycosylase and apurinic/apyrimidinic
(AP) lyase involved in base excision repair;
acts in both nucleus and mitochondrion; creates a double-strand
break at mtDNA origins that stimulates replication in response
to oxidative stress; K10773 endonuclease III [EC:4.2.99.18]
Length=399
Score = 125 bits (314), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 69/176 (39%), Positives = 97/176 (55%), Gaps = 11/176 (6%)
Query 4 KERRFAVLVAVMLSSQTKDEQT--------HACVNRLRENGLLNPYAIAACDPEKLRGLL 55
++ R VL+ VMLSSQTKDE T C++ L + A+ + KL L+
Sbjct 140 RDYRLQVLLGVMLSSQTKDEVTAMAMLNIMRYCIDELHSEEGMTLEAVLQINETKLDELI 199
Query 56 FGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGI 115
VGFH K ++ +IL + +VP T+++L+ L GVG KMA + + AW GI
Sbjct 200 HSVGFHTRKAKYILSTCKILQDQFSSDVPATINELLGLPGVGPKMAYLTLQKAWGKIEGI 259
Query 116 AVDVHVHRISNRLGWV---KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
VDVHV R++ WV K KTP +T L++ LP+ W ++N LLVGFGQ I K
Sbjct 260 CVDVHVDRLTKLWKWVDAQKCKTPDQTRTQLQNWLPKGLWTEINGLLVGFGQIITK 315
> sce:YOL043C NTG2, SCR2; DNA N-glycosylase and apurinic/apyrimidinic
(AP) lyase involved in base excision repair, localizes
to the nucleus; sumoylated; K10773 endonuclease III [EC:4.2.99.18]
Length=380
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 61/177 (34%), Positives = 89/177 (50%), Gaps = 11/177 (6%)
Query 2 DEKERRFAVLVAVMLSSQTKDEQ--------THACVNRLRENGLLNPYAIAACDPEKLRG 53
D K R L+ MLS+QT+DE+ T C+N L+ + + D L
Sbjct 143 DPKNFRLQFLIGTMLSAQTRDERMAQAALNITEYCLNTLKIAEGITLDGLLKIDEPVLAN 202
Query 54 LLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFH 113
L+ V F+ K F+K A++L+ ++P ++ ++ L GVG KM + + W
Sbjct 203 LIRCVSFYTRKANFIKRTAQLLVDNFDSDIPYDIEGILSLPGVGPKMGYLTLQKGWGLIA 262
Query 114 GIAVDVHVHRISNRLGWV---KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
GI VDVHVHR+ WV K KT + T L+ LP W ++N +LVGFGQ IC
Sbjct 263 GICVDVHVHRLCKMWNWVDPIKCKTAEHTRKELQVWLPHSLWYEINTVLVGFGQLIC 319
> eco:b1633 nth, ECK1629, JW1625; DNA glycosylase and apyrimidinic
(AP) lyase (endonuclease III) (EC:4.2.99.18); K10773 endonuclease
III [EC:4.2.99.18]
Length=211
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 49/160 (30%), Positives = 83/160 (51%), Gaps = 2/160 (1%)
Query 8 FAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNKTTF 67
F +L+AV+LS+Q D + +L P A+ E ++ + +G +N+K
Sbjct 30 FELLIAVLLSAQATDVSVNKATAKLYPVAN-TPAAMLELGVEGVKTYIKTIGLYNSKAEN 88
Query 68 LKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISNR 127
+ + IL+++H GEVP+ L L GVGRK AN+V++ A+ + IAVD H+ R+ NR
Sbjct 89 IIKTCRILLEQHNGEVPEDRAALEALPGVGRKTANVVLNTAF-GWPTIAVDTHIFRVCNR 147
Query 128 LGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
+ K ++ E L +P + D + L+ G+ C
Sbjct 148 TQFAPGKNVEQVEEKLLKVVPAEFKVDCHHWLILHGRYTC 187
> bbo:BBOV_I004290 19.m02329; 8-oxoguanine DNA glycosylase; K03660
N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18]
Length=266
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 47/84 (55%), Gaps = 2/84 (2%)
Query 43 IAACDPEKLRGLLFGVGFHNNKTTFLKE-AAEILIKKHKGEVPQTLDDLMQLRGVGRKMA 101
++A D L GL F + +FL+E E L+ P+ L++L++L GVGRK+A
Sbjct 160 LSAADFTSL-GLGFRAKYVARSISFLRENGLEWLLNLRDIPYPEALEELVKLPGVGRKVA 218
Query 102 NIVMHAAWNSFHGIAVDVHVHRIS 125
+ ++ + + VDVHV+RI+
Sbjct 219 DCILLYSLGKRECVPVDVHVNRIA 242
> dre:791194 ogg1, zgc:158858; 8-oxoguanine DNA glycosylase (EC:4.2.99.18);
K03660 N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18]
Length=391
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 46/91 (50%), Gaps = 14/91 (15%)
Query 46 CDPEKLRGLLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTL---------DDLMQLRGV 96
C +LR L FG + FL++++++++ H + Q+L D L L GV
Sbjct 219 CVEMRLRDLGFGY-----RARFLQQSSQMIMNSHHPDWLQSLRSTPYLQARDALRTLPGV 273
Query 97 GRKMANIVMHAAWNSFHGIAVDVHVHRISNR 127
G K+A+ V + + F + VD HV +I+ R
Sbjct 274 GLKVADCVCLMSLDKFEALPVDTHVWQIAKR 304
> dre:558403 mutyh, si:ch211-241d21.4; mutY homolog (E. coli);
K03575 A/G-specific adenine glycosylase [EC:3.2.2.-]
Length=526
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 45/176 (25%), Positives = 81/176 (46%), Gaps = 15/176 (8%)
Query 1 KDEKERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGF 60
+D+ R +AV V+ ++ QT+ NR + +AA E++ + G+G+
Sbjct 84 QDDNIRTYAVWVSEIMLQQTQVATVIDYYNRWMKR-WPTVEKLAAATLEEVNQMWSGLGY 142
Query 61 HNNKTTFLKEAAEILIKKHKGEVPQTLDDLM-QLRGVGRKMANIVMHAAWNSFHGIAVDV 119
++ + L E A+ ++ + G++P+T L+ QL GVGR A + A G AVD
Sbjct 143 YS-RGRRLHEGAQKVVSELDGQMPKTTAGLLKQLPGVGRYTAGAIGSIALGQVTG-AVDG 200
Query 120 HVHRISNRLGWVKTKTPQETEIALEDCLPR--------KHWEDVNLLLVGFGQQIC 167
+V R+ R V+ + A+ D L R + D N ++ G ++C
Sbjct 201 NVIRVLCR---VRAIGADSSSPAVTDALWRIADALVDPERPGDFNQAMMELGARVC 253
> tpv:TP01_0968 8-oxoguanine DNA-glycosylase; K03660 N-glycosylase/DNA
lyase [EC:3.2.2.- 4.2.99.18]
Length=279
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 53/112 (47%), Gaps = 19/112 (16%)
Query 35 NGLLNPYAIAACD------PEKLRGLLFGVGFHNNKTTFLKEAAEIL-------IKKHKG 81
N + + YA + D PE+L+ L G+G+ ++ F+ EIL + K
Sbjct 153 NEIFDFYAFPSVDQLGRATPEQLKKL--GLGY---RSEFIFRTVEILNSRGLNWLYSLKN 207
Query 82 EVPQTLDD-LMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISNRLGWVK 132
E T L L GVGRK+A+ V + + VDVH+ +I+N + VK
Sbjct 208 EDSDTCKSALTSLPGVGRKVADCVSLFSLGKRDVVPVDVHIQKIANTMFGVK 259
> ath:AT3G47830 HhH-GPD base excision DNA repair protein-related
Length=293
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 41/81 (50%), Gaps = 5/81 (6%)
Query 89 DLMQLRGVGRKMANIVMHAAWNSFHG-IAVDVHVHRISNRLGWV-KTKTPQETEIALEDC 146
+L +GVG K + V+ +N H VD HV I+ LGWV KT +T + L
Sbjct 186 ELSHFKGVGPKTVSCVL--MFNLQHNDFPVDTHVFEIAKALGWVPKTADRNKTYVHLNRK 243
Query 147 LPRKHWEDVNLLLVGFGQQIC 167
+P + D+N LL G +IC
Sbjct 244 IPDELKFDLNCLLYTHG-KIC 263
> eco:b2961 mutY, ECK2956, JW2928, micA, mutB; adenine DNA glycosylase
(EC:3.2.2.-); K03575 A/G-specific adenine glycosylase
[EC:3.2.2.-]
Length=350
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 53/119 (44%), Gaps = 7/119 (5%)
Query 54 LLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFH 113
L G+G++ L +AA+ + H G+ P+T +++ L GVGR A ++ + H
Sbjct 76 LWTGLGYYARARN-LHKAAQQVATLHGGKFPETFEEVAALPGVGRSTAGAILSLSLGK-H 133
Query 114 GIAVDVHVHRISNRL----GWVKTKTPQETEIAL-EDCLPRKHWEDVNLLLVGFGQQIC 167
+D +V R+ R GW K + +L E P E N ++ G IC
Sbjct 134 FPILDGNVKRVLARCYAVSGWPGKKEVENKLWSLSEQVTPAVGVERFNQAMMDLGAMIC 192
> ath:AT5G04560 DME; DME (DEMETER); DNA N-glycosylase/ DNA-(apurinic
or apyrimidinic site) lyase
Length=1987
Score = 35.8 bits (81), Expect = 0.070, Method: Composition-based stats.
Identities = 30/102 (29%), Positives = 49/102 (48%), Gaps = 15/102 (14%)
Query 59 GFHNNKTTFLKEAAEILIKKHKG-------EVP--QTLDDLMQLRGVGRKMANIVMHAAW 109
G +N +K+ E ++K H G E P + D L+ +RG+G K V
Sbjct 1495 GMNNMLAVRIKDFLERIVKDHGGIDLEWLRESPPDKAKDYLLSIRGLGLKSVECVRLL-- 1552
Query 110 NSFHGIA--VDVHVHRISNRLGWVKTKT-PQETEIALEDCLP 148
+ H +A VD +V RI+ R+GWV + P+ ++ L + P
Sbjct 1553 -TLHNLAFPVDTNVGRIAVRMGWVPLQPLPESLQLHLLELYP 1593
> pfa:PFI0835c N-glycosylase/DNA lyase, putative (EC:3.2.2.- 4.2.99.18);
K03660 N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18]
Length=657
Score = 35.0 bits (79), Expect = 0.12, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 43/86 (50%), Gaps = 15/86 (17%)
Query 49 EKLRGLLFGVGFHNNKTTFLKEAAEILIK----------KHKGEVPQTLDDLMQLRGVGR 98
+ LR L FG ++ ++ E+A++L+ K++ + +D L+Q G+G
Sbjct 465 DDLRSLGFGY-----RSNYVIESAKMLVNMGEEEWIENLKNEEKTKTCIDKLIQFPGIGL 519
Query 99 KMANIVMHAAWNSFHGIAVDVHVHRI 124
K+AN + N + I +D H++ I
Sbjct 520 KVANCICLFGLNKYDCIPIDTHIYDI 545
> ath:AT3G10010 DML2; DML2 (DEMETER-LIKE 2); 4 iron, 4 sulfur
cluster binding / catalytic/ endonuclease
Length=1332
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 54/119 (45%), Gaps = 15/119 (12%)
Query 42 AIAACDPEKLRGLLFGVGFHNNKTTFLKEAAEILIKKHKG-------EVP--QTLDDLMQ 92
A+ D K+ ++ G +N +K L+KKH +VP + + L+
Sbjct 819 ALRCTDVHKIANIIIKRGMNNMLAERIKAFLNRLVKKHGSIDLEWLRDVPPDKAKEYLLS 878
Query 93 LRGVGRKMANIVMHAAWNSFHGIA--VDVHVHRISNRLGWVKTKT-PQETEIALEDCLP 148
+ G+G K V S H IA VD +V RI+ RLGWV + P E ++ L + P
Sbjct 879 INGLGLKSVECVRLL---SLHQIAFPVDTNVGRIAVRLGWVPLQPLPDELQMHLLELYP 934
> ath:AT4G12740 adenine-DNA glycosylase-related / MYH-related;
K03575 A/G-specific adenine glycosylase [EC:3.2.2.-]
Length=630
Score = 33.9 bits (76), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 37/147 (25%), Positives = 68/147 (46%), Gaps = 22/147 (14%)
Query 1 KDEKERR-FAVLVAVMLSSQTKDE----------QTHACVNRLRENGLLNPYAIAACDPE 49
+ EKERR + V V+ ++ QT+ + Q + L + L N + +
Sbjct 154 ESEKERRAYEVWVSEIMLQQTRVQTVMKYYKRWMQKWPTIYDLGQASLENLIVSRSRELS 213
Query 50 KLRG--------LLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMA 101
LRG + G+G++ + FL E A++++ +G P LM+++G+G+ A
Sbjct 214 FLRGNEKKEVNEMWAGLGYYR-RARFLLEGAKMVVAGTEG-FPNQASSLMKVKGIGQYTA 271
Query 102 NIVMHAAWNSFHGIAVDVHVHRISNRL 128
+ A+N + VD +V R+ RL
Sbjct 272 GAIASIAFNEAVPV-VDGNVIRVLARL 297
> mmu:18294 Ogg1, Mmh; 8-oxoguanine DNA-glycosylase 1 (EC:4.2.99.18);
K03660 N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18]
Length=345
Score = 33.5 bits (75), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 55/110 (50%), Gaps = 21/110 (19%)
Query 35 NGLLNPYAIAACDPEK-LRGLLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDL--- 90
+G N +A+A + E LR L G+G+ + +++ +A+ ++++ G P L L
Sbjct 179 HGFPNLHALAGPEAETHLRKL--GLGY---RARYVRASAKAILEEQGG--PAWLQQLRVA 231
Query 91 ---------MQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISNR-LGW 130
L GVG K+A+ + A + + VDVHV +I++R GW
Sbjct 232 PYEEAHKALCTLPGVGAKVADCICLMALDKPQAVPVDVHVWQIAHRDYGW 281
> tgo:TGME49_113470 helix-hairpin-helix motif-containing protein
(EC:4.2.99.18)
Length=833
Score = 32.3 bits (72), Expect = 0.77, Method: Composition-based stats.
Identities = 20/80 (25%), Positives = 42/80 (52%), Gaps = 2/80 (2%)
Query 49 EKLRGLLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAA 108
E++ + G+G++ LK A+ ++++ GE+P ++ L+ + G+G + A
Sbjct 305 EEVSQMWSGLGYYRRARQLLK-GAQTVVQEFDGELPGDVEKLLSIPGIGPYTGGAISAIA 363
Query 109 WNSFHGIAVDVHVHRISNRL 128
+ + AVD +V R+ RL
Sbjct 364 FGN-RAAAVDGNVLRVLARL 382
> tgo:TGME49_116470 hypothetical protein
Length=3152
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 17/80 (21%), Positives = 34/80 (42%), Gaps = 0/80 (0%)
Query 63 NKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVH 122
+ T L+ A + ++ Q L+ L RG G+ ++ A W+ G
Sbjct 623 DATLILRRFASWIEEEGTASAAQPLECLFPQRGDGKSPESVAQRAPWDPLQGTESVWRAR 682
Query 123 RISNRLGWVKTKTPQETEIA 142
++ +GW+ + P E+E+
Sbjct 683 LLACSVGWLLWQFPSESELG 702
> dre:100319387 si:dkey-46a10.2; K10291 F-box protein 4
Length=386
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 10 VLVAVMLSSQTKDEQTHACVN---RLRENGLLNPYAIAACDPEKLRGLLFGVGF 60
VL+ +S +T D Q CV +L+ + L+NP+ + E L GLL G+ +
Sbjct 307 VLILSCVSRETPDRQRIHCVTIAHQLQLSSLVNPWMVLDTTAETLTGLLDGIDW 360
> cel:D2030.3 hypothetical protein
Length=508
Score = 30.0 bits (66), Expect = 3.5, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Query 42 AIAACDPEKLRGL---LFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVG 97
A DP +RGL LFG+ N T L E+L+ K + +DL+QLR +G
Sbjct 113 AAQTSDP-NIRGLAAELFGLRISNQNLTMLLNTLEVLLSATK--IKNVSNDLLQLRTLG 168
> cpv:cgd1_1810 hypothetical protein
Length=673
Score = 29.6 bits (65), Expect = 5.5, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 60 FHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDV 119
F NNK T +K ++ +K KGE+ + LD++ QL+ + ++ NS G+ +
Sbjct 149 FSNNKNT-IKMEYKVNLKDRKGEIKELLDNINQLKSKASQNETCIVAKLMNS-RGVYCGM 206
Query 120 H 120
H
Sbjct 207 H 207
> mmu:100041373 Gm3297; predicted gene 3297
Length=698
Score = 28.9 bits (63), Expect = 8.7, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 28/59 (47%), Gaps = 5/59 (8%)
Query 15 MLSSQTKDEQ-THAC---VNRLRENGL-LNPYAIAACDPEKLRGLLFGVGFHNNKTTFL 68
M + + EQ H C +N LRE G LN AI CDP + L+ V + T+L
Sbjct 556 MYTKPKRPEQYLHNCLIIINSLREIGFDLNIQAIDICDPNPVLMLMLCVYLYERLPTYL 614
> sce:YML060W OGG1; Mitochondrial glycosylase/lyase that specifically
excises 7,8-dihydro-8-oxoguanine residues located opposite
cytosine or thymine residues in DNA, repairs oxidative
damage to mitochondrial DNA, contributes to UVA resistance
(EC:4.2.99.18 3.2.2.-); K03660 N-glycosylase/DNA lyase [EC:3.2.2.-
4.2.99.18]
Length=376
Score = 28.5 bits (62), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 90 LMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISNR 127
LM GVG K+A+ V + + VDVHV RI+ R
Sbjct 232 LMSYNGVGPKVADCVCLMGLHMDGIVPVDVHVSRIAKR 269
Lambda K H
0.321 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4157683456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40