bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0811_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
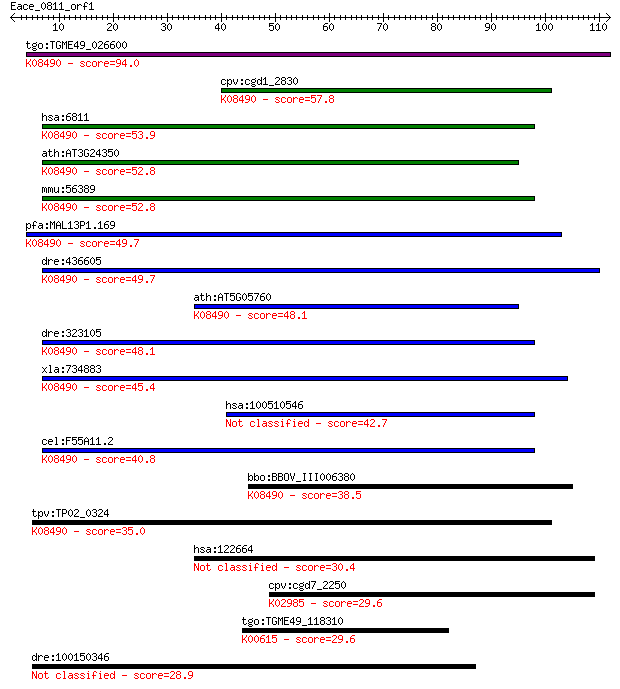
tgo:TGME49_026600 syntaxin, putative ; K08490 syntaxin 5 94.0
cpv:cgd1_2830 syntaxin 5A ortholog, possible transmembrane dom... 57.8 8e-09
hsa:6811 STX5, SED5, STX5A; syntaxin 5; K08490 syntaxin 5 53.9
ath:AT3G24350 SYP32; SYP32 (SYNTAXIN OF PLANTS 32); SNAP recep... 52.8 3e-07
mmu:56389 Stx5a, 0610031F24Rik, D19Ertd627e, Stx5; syntaxin 5A... 52.8 3e-07
pfa:MAL13P1.169 PfSyn5, PfStx5; Qa-SNARE protein, putative; K0... 49.7 2e-06
dre:436605 stx5a, wu:fj81f08, zgc:91864; syntaxin 5A; K08490 s... 49.7 3e-06
ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP recep... 48.1 6e-06
dre:323105 stx5al, wu:fb81g12, zgc:73276; syntaxin 5A, like; K... 48.1 7e-06
xla:734883 stx5, MGC114979; syntaxin 5; K08490 syntaxin 5 45.4
hsa:100510546 syntaxin-5-like 42.7 3e-04
cel:F55A11.2 syn-3; SYNtaxin family member (syn-3); K08490 syn... 40.8 0.001
bbo:BBOV_III006380 17.m07568; hypothetical protein; K08490 syn... 38.5 0.006
tpv:TP02_0324 syntaxin 5; K08490 syntaxin 5 35.0
hsa:122664 TPPP2, C14orf8, P18, p25beta; tubulin polymerizatio... 30.4 1.3
cpv:cgd7_2250 40S ribosomal protein S3, KH domain ; K02985 sma... 29.6 2.2
tgo:TGME49_118310 transketolase, putative (EC:2.2.1.3); K00615... 29.6 2.6
dre:100150346 novel immune type receptor protein-like 28.9 4.2
> tgo:TGME49_026600 syntaxin, putative ; K08490 syntaxin 5
Length=283
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 53/113 (46%), Positives = 70/113 (61%), Gaps = 9/113 (7%)
Query 4 MGYDRTEEFLAAAESFQLPGGGAQ-----NDTQRHFSDADRHFNALASEMGNALHATSLK 58
M DRT +FLA AE PG +Q + T RH AD FNA A+E+G LH TSLK
Sbjct 1 MPCDRTADFLAFAERAS-PGAISQARELRSRTVRH---ADNSFNASAAEIGTQLHRTSLK 56
Query 59 LQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFARAVENAGGGG 111
L+EL K ARQ IYN +T Q +LT++I+K+I LNCK+D + +++G G
Sbjct 57 LKELAKFARQRSIYNDRTAQTQDLTYEIKKSITELNCKIDYLEQIAKDSGSEG 109
> cpv:cgd1_2830 syntaxin 5A ortholog, possible transmembrane domain
or GPI at C-terminus ; K08490 syntaxin 5
Length=329
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 40 HFNALASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDV 99
FN LASE+ +++TSLK++EL ++ +Q G++ +T QI +LT DI+ ++ LN +++V
Sbjct 48 QFNLLASEISQEMNSTSLKIEELNRIVKQKGLFRDRTNQIHQLTEDIKTSVTELNSRLEV 107
Query 100 F 100
Sbjct 108 L 108
> hsa:6811 STX5, SED5, STX5A; syntaxin 5; K08490 syntaxin 5
Length=355
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/92 (35%), Positives = 54/92 (58%), Gaps = 1/92 (1%)
Query 7 DRTEEFLAAAESFQLPGGGAQNDTQRHFSDADR-HFNALASEMGNALHATSLKLQELGKL 65
DRT+EFL+A +S Q G Q + + R F +A +G L T KL++L L
Sbjct 59 DRTQEFLSACKSLQTRQNGIQTNKPALRAVRQRSEFTLMAKRIGKDLSNTFAKLEKLTIL 118
Query 66 ARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
A++ +++ K V+I ELT+ I++ IN+LN ++
Sbjct 119 AKRKSLFDDKAVEIEELTYIIKQDINSLNKQI 150
> ath:AT3G24350 SYP32; SYP32 (SYNTAXIN OF PLANTS 32); SNAP receptor;
K08490 syntaxin 5
Length=347
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 34/100 (34%), Positives = 55/100 (55%), Gaps = 12/100 (12%)
Query 7 DRTEEFLAAAESFQ-----------LPGGGAQNDTQRHFSDADR-HFNALASEMGNALHA 54
DR++EF E+ + +P G +ND R ++ FN AS +G A++
Sbjct 12 DRSDEFFKIVETLRRSIAPAPAANNVPYGNNRNDGARREDLINKSEFNKRASHIGLAINQ 71
Query 55 TSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALN 94
TS KL +L KLA++ +++ T +I ELT I++ I+ALN
Sbjct 72 TSQKLSKLAKLAKRTSVFDDPTQEIQELTVVIKQEISALN 111
> mmu:56389 Stx5a, 0610031F24Rik, D19Ertd627e, Stx5; syntaxin
5A; K08490 syntaxin 5
Length=355
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 32/92 (34%), Positives = 52/92 (56%), Gaps = 1/92 (1%)
Query 7 DRTEEFLAAAESFQLPGGGAQNDTQR-HFSDADRHFNALASEMGNALHATSLKLQELGKL 65
DRT+EF +A +S Q G Q H + F +A +G L T KL++L L
Sbjct 59 DRTQEFQSACKSLQSRQNGIQTSKPALHAARQCSEFTLMARRIGKDLSNTFAKLEKLTIL 118
Query 66 ARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
A++ +++ K V+I ELT+ I++ IN+LN ++
Sbjct 119 AKRKSLFDDKAVEIEELTYIIKQDINSLNKQI 150
> pfa:MAL13P1.169 PfSyn5, PfStx5; Qa-SNARE protein, putative;
K08490 syntaxin 5
Length=281
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 53/100 (53%), Gaps = 4/100 (4%)
Query 4 MGY-DRTEEFLAAAESFQLPGGGAQNDTQRHFSDADRHFNALASEMGNALHATSLKLQEL 62
M Y D+TEEF E +L R D LAS++ + L + KLQ+L
Sbjct 1 MPYVDKTEEFFKIIE--KLSNDNINIRKNRSIV-QDTQVGELASKITDLLQSGYQKLQQL 57
Query 63 GKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFAR 102
+ +Q GI+N KT +I ELT+++++TI + ++D+ +
Sbjct 58 ERCVKQKGIFNDKTSEIEELTYEVKQTITDVTNELDLLVQ 97
> dre:436605 stx5a, wu:fj81f08, zgc:91864; syntaxin 5A; K08490
syntaxin 5
Length=302
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 57/109 (52%), Gaps = 10/109 (9%)
Query 7 DRTEEFLAAAESFQLPGGGAQNDTQRHFSDADR------HFNALASEMGNALHATSLKLQ 60
DRT EF +A +S Q G QN T H A+ F +A +G L T KL+
Sbjct 5 DRTLEFQSACKSLQ--GRQLQNGT--HSKPANNALKQRSDFTLMAKRIGKDLSNTFAKLE 60
Query 61 ELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFARAVENAGG 109
+L LA++ +++ K V+I ELT+ I++ IN+LN ++ V + G
Sbjct 61 KLTILAKRKSLFDDKAVEIEELTYIIKQDINSLNKQIAQLQDLVRSRSG 109
> ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP receptor;
K08490 syntaxin 5
Length=336
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/60 (45%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 35 SDADRHFNALASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALN 94
S FN AS +G + TS K+ L KLA+Q I+N +TV+I ELT IR I LN
Sbjct 41 SSPGSEFNKKASRIGLGIKETSQKITRLAKLAKQSTIFNDRTVEIQELTVLIRNDITGLN 100
> dre:323105 stx5al, wu:fb81g12, zgc:73276; syntaxin 5A, like;
K08490 syntaxin 5
Length=298
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 53/96 (55%), Gaps = 9/96 (9%)
Query 7 DRTEEFLAAAESFQLPGGGAQ-----NDTQRHFSDADRHFNALASEMGNALHATSLKLQE 61
DRT EF + +S Q GAQ N+ + SD F LA +G L T KL++
Sbjct 5 DRTGEFQSVCKSLQGRQNGAQPVRAVNNAIQKRSD----FTLLAKRIGRDLSNTFAKLEK 60
Query 62 LGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
L LA++ +++ K +I ELT+ +++ IN+LN ++
Sbjct 61 LTILAKRKSLFDDKATEIDELTYIVKQDINSLNKQI 96
> xla:734883 stx5, MGC114979; syntaxin 5; K08490 syntaxin 5
Length=298
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 53/101 (52%), Gaps = 4/101 (3%)
Query 7 DRTEEFLAAAESFQLPGGGAQNDTQRHFSDADR-HFNALASEMGNALHATSLKLQELGKL 65
DRT EF++ +S Q G Q + + R F +A +G L T KL++L L
Sbjct 5 DRTAEFISTCKSLQGRQNGVQLSSPSLNAVKQRSEFTLMAKRIGKDLSNTFSKLEKLTIL 64
Query 66 ARQCGIYNVKTVQIPELTFDIRKTINALN---CKVDVFARA 103
A++ +++ K +I ELT+ I++ I +LN ++ F RA
Sbjct 65 AKRKSLFDDKAAEIEELTYIIKQDIGSLNQQIAQLQSFVRA 105
> hsa:100510546 syntaxin-5-like
Length=295
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 41 FNALASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
F +A +G L T KL++L LA++ +++ K V+I ELT+ I++ IN+LN ++
Sbjct 34 FTLMAKRIGKDLSNTFAKLEKLTILAKRKSLFDDKAVEIEELTYIIKQDINSLNKQI 90
> cel:F55A11.2 syn-3; SYNtaxin family member (syn-3); K08490 syntaxin
5
Length=413
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 46/93 (49%), Gaps = 3/93 (3%)
Query 7 DRTEEFLAAAESFQLPGG--GAQNDTQRHFSDADRHFNALASEMGNALHATSLKLQELGK 64
DRT EF A A+S+++ G + + FN LA +G L T K+++L +
Sbjct 111 DRTSEFRATAKSYEMKAAANGIRPQPKHEMLSESVQFNQLAKRIGKELSQTCAKMEKLAE 170
Query 65 LARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
A++ Y ++ QI L+ ++ I LN ++
Sbjct 171 YAKKKSCYEERS-QIDHLSSIVKSDITGLNKQI 202
> bbo:BBOV_III006380 17.m07568; hypothetical protein; K08490 syntaxin
5
Length=256
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 45 ASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFARAV 104
A +G L KL EL LAR+ IY T +I LT D+++ I A + K+D F V
Sbjct 40 AQRVGLQLSKCETKLTELSALARKRSIYVDHTAEIERLTNDVKEGITAASSKIDEFETKV 99
> tpv:TP02_0324 syntaxin 5; K08490 syntaxin 5
Length=285
Score = 35.0 bits (79), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 5/96 (5%)
Query 5 GYDRTEEFLAAAESFQLPGGGAQNDTQRHFSDADRHFNALASEMGNALHATSLKLQELGK 64
DRT+ F+ Q+ N+TQ + + FN A + + KL EL +
Sbjct 4 SVDRTKFFMT-----QVNQNSNLNNTQAPKTFENLEFNTDADSIYKEIEKARAKLNELSQ 58
Query 65 LARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVF 100
LA++ +Y T I LT +I+ + +D+F
Sbjct 59 LAKKRSLYLDNTSSIERLTSEIKSILTYTTSSIDLF 94
> hsa:122664 TPPP2, C14orf8, P18, p25beta; tubulin polymerization-promoting
protein family member 2
Length=170
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 35/76 (46%), Gaps = 2/76 (2%)
Query 35 SDADRHFNALASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIP--ELTFDIRKTINA 92
S+A++ F+ A+ ++ T + + KL + CGI + KTV ++ F K NA
Sbjct 3 SEAEKTFHRFAAFGESSSSGTEMNNKNFSKLCKDCGIMDGKTVTSTDVDIVFSKVKAKNA 62
Query 93 LNCKVDVFARAVENAG 108
F AV+ G
Sbjct 63 RTITFQQFKEAVKELG 78
> cpv:cgd7_2250 40S ribosomal protein S3, KH domain ; K02985 small
subunit ribosomal protein S3e
Length=221
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 30/60 (50%), Gaps = 0/60 (0%)
Query 49 GNALHATSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFARAVENAG 108
G + T +K + + + R + VK +I ELT I+K N V++FA +EN G
Sbjct 37 GVEVRVTPIKTEIIIRATRTREVLGVKGRRIRELTSLIQKRFNFPEGSVELFAERIENRG 96
> tgo:TGME49_118310 transketolase, putative (EC:2.2.1.3); K00615
transketolase [EC:2.2.1.1]
Length=699
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 44 LASEMGNA-LHATSLKLQELGKLARQCGIYNVKTVQIPE 81
L+S+ G A +H + L +EL + QCG+ + +QIPE
Sbjct 282 LSSKAGTAKVHGSPLSEEELRAVKEQCGLSPDEKLQIPE 320
> dre:100150346 novel immune type receptor protein-like
Length=259
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/82 (24%), Positives = 37/82 (45%), Gaps = 2/82 (2%)
Query 5 GYDRTEEFLAAAESFQLPGGGAQNDTQRHFSDADRHFNALASEMGNALHATSLKLQELGK 64
G + T F ++ E+ +LP A +D + +RH + G +L ++ + L
Sbjct 24 GAEETHVFFSSGENVRLPCNNALSDCTSTVWNYNRHSETVELVAGGSLKTDIMRRERLS- 82
Query 65 LARQCGIYNVKTVQIPELTFDI 86
L C + N+K V+ + F I
Sbjct 83 LGSDCSL-NIKKVKKEDYGFYI 103
Lambda K H
0.317 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2062416360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40