bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0819_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
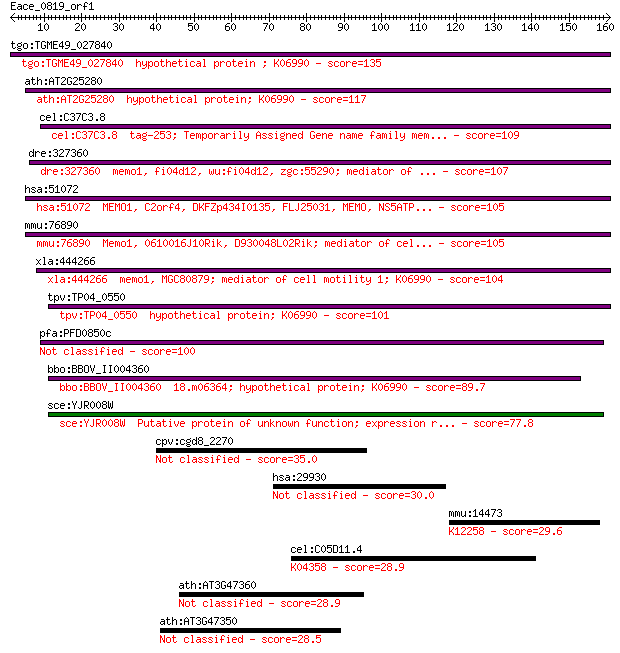
tgo:TGME49_027840 hypothetical protein ; K06990 135 4e-32
ath:AT2G25280 hypothetical protein; K06990 117 1e-26
cel:C37C3.8 tag-253; Temporarily Assigned Gene name family mem... 109 4e-24
dre:327360 memo1, fi04d12, wu:fi04d12, zgc:55290; mediator of ... 107 1e-23
hsa:51072 MEMO1, C2orf4, DKFZp434I0135, FLJ25031, MEMO, NS5ATP... 105 5e-23
mmu:76890 Memo1, 0610016J10Rik, D930048L02Rik; mediator of cel... 105 5e-23
xla:444266 memo1, MGC80879; mediator of cell motility 1; K06990 104 1e-22
tpv:TP04_0550 hypothetical protein; K06990 101 9e-22
pfa:PFD0850c Memo-like protein 100 2e-21
bbo:BBOV_II004360 18.m06364; hypothetical protein; K06990 89.7 4e-18
sce:YJR008W Putative protein of unknown function; expression r... 77.8 1e-14
cpv:cgd8_2270 hypothetical protein 35.0 0.11
hsa:29930 PCDHB1, MGC138301, MGC138303, PCDH-BETA1; protocadhe... 30.0 3.0
mmu:14473 Gc, DBP, VDB; group specific component; K12258 vitam... 29.6 4.6
cel:C05D11.4 let-756; LEThal family member (let-756); K04358 f... 28.9 7.0
ath:AT3G47360 AtHSD3; AtHSD3 (hydroxysteroid dehydrogenase 3);... 28.9 7.5
ath:AT3G47350 AtHSD2; short-chain dehydrogenase/reductase (SDR... 28.5 9.1
> tgo:TGME49_027840 hypothetical protein ; K06990
Length=302
Score = 135 bits (341), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 69/161 (42%), Positives = 94/161 (58%), Gaps = 2/161 (1%)
Query 1 LPSESAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF-SE 59
LP+ S Y TPLGD+ LDT L+ LR F ++ +DEEEHSIE+ LPF+RH +
Sbjct 86 LPASSVRAYATPLGDISLDTAVLETLRSAKLFETLALRDDEEEHSIEMQLPFLRHILRGK 145
Query 60 GVKIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPG 119
+VP++VGDL+ A L YFLQ+ NLF+ SSDFCHWG R+ Y YL
Sbjct 146 SFTLVPIVVGDLRP-SGHAAVAKALRPYFLQEGNLFVFSSDFCHWGRRFRYSYLPPATAS 204
Query 120 VPIHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
+PI I +D++ I + GF +Y E TG ++CG +
Sbjct 205 LPIFERIGILDKEGAALIEQQDPAGFQEYYERTGNTICGHN 245
> ath:AT2G25280 hypothetical protein; K06990
Length=291
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 60/157 (38%), Positives = 91/157 (57%), Gaps = 4/157 (2%)
Query 5 SAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF-SEGVKI 63
+A VY TP+G+LP+D + ++ +R G F + DE EHS+E+ LP++ F VK+
Sbjct 90 TATVYKTPIGNLPVDVEMIKEIRAMGKFGMMDLRVDEAEHSMEMHLPYLAKVFEGNNVKV 149
Query 64 VPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIH 123
VP++VG + S E+ Y +LL Y +N F VSSDFCHWG R++Y + ++ IH
Sbjct 150 VPILVGAV-SPENEAMYGELLAKYVDDPKNFFSVSSDFCHWGSRFNYMHYDNTHGA--IH 206
Query 124 AAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
+IE +D+K + I F YL ++CGRH
Sbjct 207 KSIEALDKKGMDIIETGDPDAFKKYLLEFENTICGRH 243
> cel:C37C3.8 tag-253; Temporarily Assigned Gene name family member
(tag-253); K06990
Length=350
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 60/153 (39%), Positives = 84/153 (54%), Gaps = 3/153 (1%)
Query 9 YLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF-SEGVKIVPMI 67
Y TPLGDL +D + LR T F ++E EHSIE+ LPF+ S+ IVP++
Sbjct 147 YRTPLGDLIVDHKINEELRATRHFDLMDRRDEESEHSIEMQLPFIAKVMGSKRYTIVPVL 206
Query 68 VGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHAAIE 127
VG L Q Y ++ +Y NLF++SSDFCHWG R+S F D +PI+ I
Sbjct 207 VGSLPGSRQ-QTYGNIFAHYMEDPRNLFVISSDFCHWGERFS-FSPYDRHSSIPIYEQIT 264
Query 128 KMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
MD++ + I F DYL+ T ++CGR+
Sbjct 265 NMDKQGMSAIETLNPAAFNDYLKKTQNTICGRN 297
> dre:327360 memo1, fi04d12, wu:fi04d12, zgc:55290; mediator of
cell motility 1; K06990
Length=297
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 61/158 (38%), Positives = 86/158 (54%), Gaps = 6/158 (3%)
Query 6 AGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF---SEGVK 62
A VY TPL DL +D L TG F S+ DE+EHSIE+ LP+ +
Sbjct 93 AEVYRTPLYDLRIDQKVYADLWKTGMFERMSLQTDEDEHSIEMHLPYTAKAMENHKDEFS 152
Query 63 IVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPI 122
IVP++VG L ++ Q+Y LL Y NLFI+S DFCHWG R+ Y Y ++ Q I
Sbjct 153 IVPVLVGALSGSKE-QEYGKLLSKYLADPSNLFIISPDFCHWGQRFRYTYYDESQG--EI 209
Query 123 HAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
+ +IE +D+ + I + F +YL+ ++CGRH
Sbjct 210 YRSIEHLDKMGMGIIEQLDPISFSNYLKKYHNTICGRH 247
> hsa:51072 MEMO1, C2orf4, DKFZp434I0135, FLJ25031, MEMO, NS5ATP7;
mediator of cell motility 1; K06990
Length=274
Score = 105 bits (263), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 58/159 (36%), Positives = 86/159 (54%), Gaps = 6/159 (3%)
Query 5 SAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF---SEGV 61
S +Y TPL DL +D L TG F S+ DE+EHSIE+ LP+ +
Sbjct 69 SVDIYRTPLYDLRIDQKIYGELWKTGMFERMSLQTDEDEHSIEMHLPYTAKAMESHKDEF 128
Query 62 KIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVP 121
I+P++VG L ++ Q++ L Y NLF+VSSDFCHWG R+ Y Y ++ Q
Sbjct 129 TIIPVLVGALSESKE-QEFGKLFSKYLADPSNLFVVSSDFCHWGQRFRYSYYDESQG--E 185
Query 122 IHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
I+ +IE +D+ + I + F +YL+ ++CGRH
Sbjct 186 IYRSIEHLDKMGMSIIEQLDPVSFSNYLKKYHNTICGRH 224
> mmu:76890 Memo1, 0610016J10Rik, D930048L02Rik; mediator of cell
motility 1; K06990
Length=297
Score = 105 bits (263), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 58/159 (36%), Positives = 86/159 (54%), Gaps = 6/159 (3%)
Query 5 SAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF---SEGV 61
S +Y TPL DL +D L TG F S+ DE+EHSIE+ LP+ +
Sbjct 92 SVDIYRTPLYDLRIDQKIYGELWKTGMFERMSLQTDEDEHSIEMHLPYTAKAMESHKDEF 151
Query 62 KIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVP 121
I+P++VG L ++ Q++ L Y NLF+VSSDFCHWG R+ Y Y ++ Q
Sbjct 152 TIIPVLVGALSESKE-QEFGKLFSKYLADPSNLFVVSSDFCHWGQRFRYSYYDESQG--E 208
Query 122 IHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
I+ +IE +D+ + I + F +YL+ ++CGRH
Sbjct 209 IYRSIEHLDKMGMSIIEQLDPVSFSNYLKKYHNTICGRH 247
> xla:444266 memo1, MGC80879; mediator of cell motility 1; K06990
Length=297
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/156 (35%), Positives = 86/156 (55%), Gaps = 6/156 (3%)
Query 8 VYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF---SEGVKIV 64
+Y TPL DL +D L TG F S+ DE+EHSIE+ LP+ + + IV
Sbjct 95 IYRTPLYDLHVDQKVYGELWKTGMFERMSLQTDEDEHSIEMHLPYTAKAMESHKDDLTIV 154
Query 65 PMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHA 124
P++VG L ++ Q++ + Y NLF++SSDFCHWG R+ Y Y ++ Q I+
Sbjct 155 PVLVGALSESKE-QEFGKVFSKYLADPTNLFVISSDFCHWGQRFRYTYYDESQG--EIYR 211
Query 125 AIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
+IE +D+ + I + F +YL+ ++CGRH
Sbjct 212 SIENLDKMGMSIIEQLDPVQFSNYLKKYHNTICGRH 247
> tpv:TP04_0550 hypothetical protein; K06990
Length=290
Score = 101 bits (252), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 56/152 (36%), Positives = 83/152 (54%), Gaps = 4/152 (2%)
Query 11 TPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTFS-EGVKIVPMIVG 69
TPLG L +D D + L F + E+EHSIE+ LP +R F E VK+VP++VG
Sbjct 93 TPLGPLQVDVDIVDKLSNLKGFSVINNEASEDEHSIEMHLPLLRFVFKKEPVKVVPIMVG 152
Query 70 DL-KSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHAAIEK 128
D +SL D + L+ YF + LF+ SSDFCH+G R+ + P++ IE
Sbjct 153 DFSESLAD--ELTSALVPYFNDERTLFVFSSDFCHFGSRFQFSITGYESENKPLYEKIEM 210
Query 129 MDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
+D++ + I H+ F YL T ++CGR+
Sbjct 211 LDKRGIDLIVNHKHDDFLWYLTETENTICGRN 242
> pfa:PFD0850c Memo-like protein
Length=296
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 56/155 (36%), Positives = 82/155 (52%), Gaps = 5/155 (3%)
Query 9 YLTPLGDLPLDTDTLQALRLTGAFCEASI---SEDEEEHSIELMLPFVRHTFSEG-VKIV 64
Y TP G L ++ + + + S +DEEEHSIE+ LP +++ + +KIV
Sbjct 93 YETPFGFLQINKQIISDIIKSDTHNLYSFIDPDDDEEEHSIEMQLPLIKYIIKDKDIKIV 152
Query 65 PMIVGDL-KSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIH 123
P+ VG + L+ +A+ L YF NLF+ SSDFCH+GPR+ + + I
Sbjct 153 PIYVGSIGNDLKKIDLFANPLKKYFQDQHNLFLFSSDFCHYGPRFRFTNILQKYSDTFIF 212
Query 124 AAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCG 158
IE MD+ AV I+ H GF DYL T ++CG
Sbjct 213 KQIENMDKDAVNIISHHDLTGFVDYLNETHNTICG 247
> bbo:BBOV_II004360 18.m06364; hypothetical protein; K06990
Length=245
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 50/146 (34%), Positives = 79/146 (54%), Gaps = 5/146 (3%)
Query 11 TPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTFSEG----VKIVPM 66
TP G+L +D D L F E S EEEHSIE+ LP + + ++ +K+VP+
Sbjct 70 TPFGELQVDNDITTELLKGKCFKELSKRNSEEEHSIEMQLPILHYVANKSNADHIKVVPI 129
Query 67 IVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHAAI 126
+VG + + E + L+ YF +++ +F++SSDFCH+G R+ + +PI AI
Sbjct 130 VVGYMLN-EGLEDVGQALLPYFEKEDTIFVISSDFCHFGKRFGFTRTGFEDQDMPIWKAI 188
Query 127 EKMDRKAVKFITEHQGQGFYDYLEST 152
E +D VK I EH + Y++ T
Sbjct 189 ESLDLDGVKLIVEHDLEVSNKYIKIT 214
> sce:YJR008W Putative protein of unknown function; expression
repressed by inosine and choline in an Opi1p-dependent manner;
expression induced by mild heat-stress on a non-fermentable
carbon source; K06990
Length=338
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 62/189 (32%), Positives = 84/189 (44%), Gaps = 42/189 (22%)
Query 11 TPLGDLPLDTDTLQAL-------RLTGAFCEASISEDEEEHSIELMLPFVRHTFS----- 58
TPLG+L +DTD + L F D EHS+E+ LP + T
Sbjct 98 TPLGNLKVDTDLCKTLIQKEYPENGKKLFKPMDHDTDMAEHSLEMQLPMLVETLKWREIS 157
Query 59 -EGVKIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYF------ 111
+ VK+ PM+V S++ + ++L Y NLFIVSSDFCHWG R+ Y
Sbjct 158 LDTVKVFPMMVSH-NSVDVDRCIGNILSEYIKDPNNLFIVSSDFCHWGRRFQYTGYVGSK 216
Query 112 -YLEDP-----------------QPGVPIHAAIEKMDRKAVKFITEHQGQGFYD----YL 149
L D VPI +IE MDR A+K +++ YD YL
Sbjct 217 EELNDAIQEETEVEMLTARSKLSHHQVPIWQSIEIMDRYAMKTLSDTPNGERYDAWKQYL 276
Query 150 ESTGLSVCG 158
E TG ++CG
Sbjct 277 EITGNTICG 285
> cpv:cgd8_2270 hypothetical protein
Length=1063
Score = 35.0 bits (79), Expect = 0.11, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 36/63 (57%), Gaps = 7/63 (11%)
Query 40 DEEEHSIELMLPF-----VRHTFSEGVKIVPMIVGDLKSLEDCQKYAD--LLINYFLQDE 92
DE++HSI ++LP ++ KI+P+I ++ S D ++Y D +L N ++DE
Sbjct 864 DEDDHSIPVILPKRLEGKIKDVLENEDKIIPVIPKNISSFNDKEQYNDDIILNNVDIEDE 923
Query 93 NLF 95
+ F
Sbjct 924 SEF 926
> hsa:29930 PCDHB1, MGC138301, MGC138303, PCDH-BETA1; protocadherin
beta 1
Length=818
Score = 30.0 bits (66), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 71 LKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDP 116
L+S +D + L NY L F + + FC GP+Y+ L P
Sbjct 154 LQSAQDLDVGLNGLQNYTLSANGYFHLHTRFCSHGPKYAELVLNKP 199
> mmu:14473 Gc, DBP, VDB; group specific component; K12258 vitamin
D-binding protein
Length=476
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 18/40 (45%), Gaps = 0/40 (0%)
Query 118 PGVPIHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVC 157
PG P E ++RK HQ Q F Y+E T +C
Sbjct 106 PGTPECCTKEGLERKLCMAALSHQPQEFPTYVEPTNDEIC 145
> cel:C05D11.4 let-756; LEThal family member (let-756); K04358
fibroblast growth factor
Length=425
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 31/65 (47%), Gaps = 8/65 (12%)
Query 76 DCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHAAIEKMDRKAVK 135
+C +++ NY+ NL+ C +G R++ +Y+E + G P K RKA
Sbjct 154 ECVFLEEMMENYY----NLYAS----CAYGDRFNPWYIELRRSGKPRRGPNSKKRRKASH 205
Query 136 FITEH 140
F+ H
Sbjct 206 FLVVH 210
> ath:AT3G47360 AtHSD3; AtHSD3 (hydroxysteroid dehydrogenase 3);
binding / catalytic/ oxidoreductase
Length=309
Score = 28.9 bits (63), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 29/49 (59%), Gaps = 3/49 (6%)
Query 46 IELMLPFVRHTFSEGVKIVPMIVGDLKSLEDCQKYADLLINYFLQDENL 94
+E++ R S V I+P GD+ ++EDC+K+ D I +F + ++L
Sbjct 84 LEIVAETSRQLGSGNVIIIP---GDVSNVEDCKKFIDETIRHFGKLDHL 129
> ath:AT3G47350 AtHSD2; short-chain dehydrogenase/reductase (SDR)
family protein
Length=321
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 3/48 (6%)
Query 41 EEEHSIELMLPFVRHTFSEGVKIVPMIVGDLKSLEDCQKYADLLINYF 88
+ +E++ R S V I+P GD+ ++EDC+K+ D I++F
Sbjct 78 RRKDRLEIVAETSRQLGSGDVIIIP---GDVSNVEDCKKFIDETIHHF 122
Lambda K H
0.321 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40