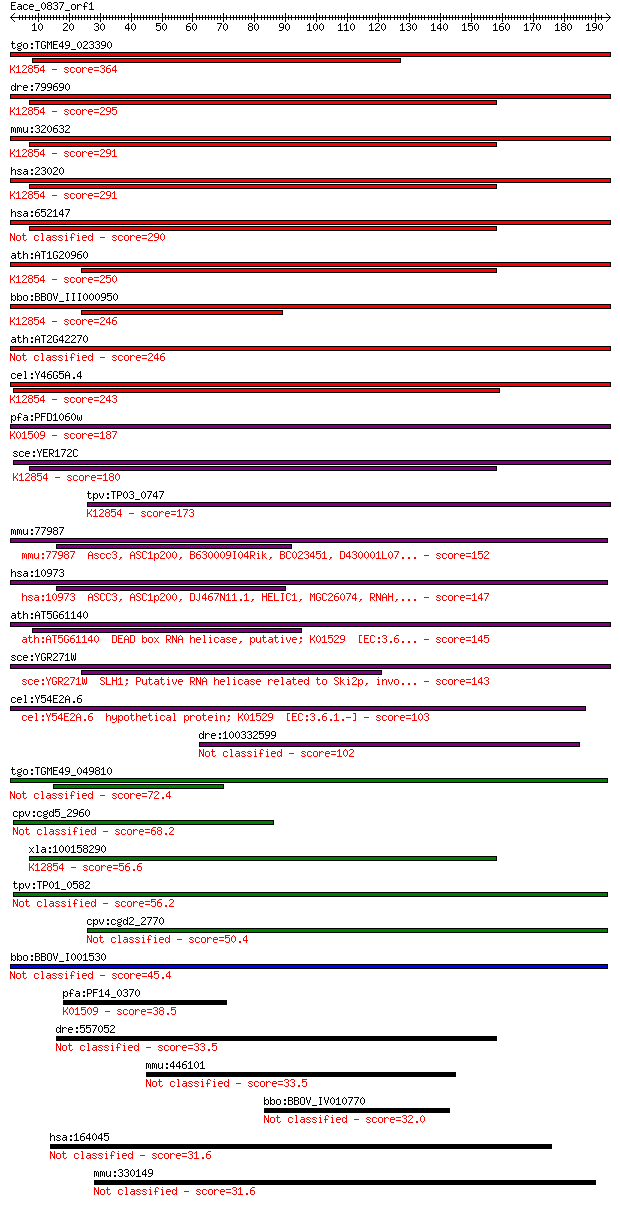
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0837_orf1
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 364 8e-101
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 295 8e-80
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 291 1e-78
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 291 1e-78
hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase... 290 2e-78
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 250 2e-66
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 246 3e-65
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 246 4e-65
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 243 2e-64
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 187 2e-47
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 180 3e-45
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 173 3e-43
mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07... 152 5e-37
hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,... 147 2e-35
ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6... 145 1e-34
sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, invo... 143 4e-34
cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-] 103 3e-22
dre:100332599 mutagen-sensitive 308-like 102 9e-22
tgo:TGME49_049810 activating signal cointegrator 1 complex sub... 72.4 1e-12
cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the sec... 68.2 2e-11
xla:100158290 snrnp200, ascc3l1; small nuclear ribonucleoprote... 56.6 6e-08
tpv:TP01_0582 RNA helicase 56.2 7e-08
cpv:cgd2_2770 U5 small nuclear ribonucleoprotein 200kDA helica... 50.4 4e-06
bbo:BBOV_I001530 19.m02171; helicase 45.4 1e-04
pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosi... 38.5 0.015
dre:557052 hfm1, si:ch211-83f14.5; HFM1, ATP-dependent DNA hel... 33.5 0.41
mmu:446101 Xrra1, AI449753; X-ray radiation resistance associa... 33.5 0.45
bbo:BBOV_IV010770 23.m05766; hypothetical protein 32.0 1.5
hsa:164045 HFM1, FLJ36760, FLJ39011, MER3, MGC163397, RP11-539... 31.6 1.6
mmu:330149 Hfm1, A330009G12Rik, Gm1046, Sec63d1; HFM1, ATP-dep... 31.6 2.0
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 364 bits (935), Expect = 8e-101, Method: Compositional matrix adjust.
Identities = 171/194 (88%), Positives = 185/194 (95%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEK+ELQRLMERVPIPVKGSPDE SSKVNVLLQAYISKLKLEG AMMADMVYVQQSANR
Sbjct 1085 EEEKVELQRLMERVPIPVKGSPDETSSKVNVLLQAYISKLKLEGLAMMADMVYVQQSANR 1144
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
IMRAIF+ICLRRGWA LA+RAL+ C EI RMWS+MTPLRQFKVLPEEL+RKIEKKDLPF
Sbjct 1145 IMRAIFEICLRRGWAMLALRALRFCKEIDRRMWSSMTPLRQFKVLPEELLRKIEKKDLPF 1204
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+RYYDL+STEIGELVRVPKMGKLLHRLIH FPKLELAAFVQPL+R+CLVVELTITPDFQW
Sbjct 1205 ERYYDLSSTEIGELVRVPKMGKLLHRLIHQFPKLELAAFVQPLTRTCLVVELTITPDFQW 1264
Query 181 DQKIHGNGEVFWIL 194
+ K+HG+GEVFW+L
Sbjct 1265 EAKVHGSGEVFWVL 1278
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 63/119 (52%), Gaps = 4/119 (3%)
Query 8 QRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFD 67
QRL R+P + ++PS+K +LL A+ ++ L ++AD + + + R++ A+ D
Sbjct 1936 QRLGVRLPANSE-DLNKPSTKALILLYAHFNRTPLPS-DLIADQKVLLEPSIRLLHALVD 1993
Query 68 ICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYYDL 126
+ GW A+ A+++C + M + + L+Q +EL+ + K++ D +DL
Sbjct 1994 VISSNGWLVPALSAMEICQAVVQAMTTACSALKQLPHFTDELVE--QAKEMGVDDIFDL 2050
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 295 bits (754), Expect = 8e-80, Method: Compositional matrix adjust.
Identities = 135/194 (69%), Positives = 163/194 (84%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEKLELQ+L+ERVPIPVK S +EPS+K+NVLLQAYIS+LKLEGFA+MADMVYV QSA R
Sbjct 1028 EEEKLELQKLLERVPIPVKESIEEPSAKINVLLQAYISQLKLEGFALMADMVYVTQSAGR 1087
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+MRAIF+I L RGWAQL + L LC I RMW +M+PLRQF+ LPEE+++KIEKK+ PF
Sbjct 1088 LMRAIFEIVLSRGWAQLTDKTLNLCKMIDKRMWQSMSPLRQFRKLPEEVIKKIEKKNFPF 1147
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+R YDL EIGEL+R+PKMGK +H+ +H FPKL+LA +QP++RS L VELTITPDFQW
Sbjct 1148 ERLYDLNHNEIGELIRMPKMGKTIHKYVHQFPKLDLAVHLQPITRSTLKVELTITPDFQW 1207
Query 181 DQKIHGNGEVFWIL 194
D KIHG+ E FWIL
Sbjct 1208 DDKIHGSSEAFWIL 1221
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/156 (24%), Positives = 80/156 (51%), Gaps = 8/156 (5%)
Query 7 LQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAI 65
L++L ++VP + ++P K N+LLQA++S+++L + +D + A R+++A
Sbjct 1864 LRQLAQKVPHKLNNPKFNDPHVKTNLLLQAHLSRMQLSA-ELQSDTEEILSKAVRLIQAC 1922
Query 66 FDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYYD 125
D+ GW A+ A++L + MWS + LRQ EL+++ K + + +D
Sbjct 1923 VDVLSSNGWLSPALAAMELAQMVTQAMWSKDSYLRQLPHFTSELIKRCTDKGV--ESIFD 1980
Query 126 LTSTEIGELVRVPKMGKL----LHRLIHSFPKLELA 157
+ E + + ++ + + R + +P +EL+
Sbjct 1981 IMEMEDEDRTGLLQLSDVQVADVARFCNRYPNIELS 2016
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 291 bits (745), Expect = 1e-78, Method: Compositional matrix adjust.
Identities = 133/194 (68%), Positives = 163/194 (84%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEKLELQ+L+ERVPIPVK S +EPS+K+NVLLQA+IS+LKLEGFA+MADMVYV QSA R
Sbjct 1031 EEEKLELQKLLERVPIPVKESIEEPSAKINVLLQAFISQLKLEGFALMADMVYVTQSAGR 1090
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+MRAIF+I L RGWAQL + L LC I RMW +M PLRQF+ LPEE+++KIEKK+ PF
Sbjct 1091 LMRAIFEIVLNRGWAQLTDKTLNLCKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPF 1150
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+R YDL EIGEL+R+PKMGK +H+ +H FPKLEL+ +QP++RS L VELTITPDFQW
Sbjct 1151 ERLYDLNHNEIGELIRMPKMGKTIHKYVHLFPKLELSVHLQPITRSTLKVELTITPDFQW 1210
Query 181 DQKIHGNGEVFWIL 194
D+K+HG+ E FWIL
Sbjct 1211 DEKVHGSSEAFWIL 1224
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 38/164 (23%), Positives = 78/164 (47%), Gaps = 24/164 (14%)
Query 7 LQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAI 65
L++L ++VP + ++P K N+LLQA++S+++L + +D + A R+++A
Sbjct 1868 LRQLAQKVPHKLNNPKFNDPHVKTNLLLQAHLSRMQLSA-ELQSDTEEILSKAIRLIQAC 1926
Query 66 FDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRK------------I 113
D+ GW A+ A++L + MWS + L+Q E +++ +
Sbjct 1927 VDVLSSNGWLSPALAAMELAQMVTQAMWSKDSYLKQLPHFTSEHIKRCTDKGVESVFDIM 1986
Query 114 EKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELA 157
E +D + LT ++I ++ R + +P +EL+
Sbjct 1987 EMEDEERNALLQLTDSQIADVA----------RFCNRYPNIELS 2020
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 291 bits (745), Expect = 1e-78, Method: Compositional matrix adjust.
Identities = 133/194 (68%), Positives = 163/194 (84%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEKLELQ+L+ERVPIPVK S +EPS+K+NVLLQA+IS+LKLEGFA+MADMVYV QSA R
Sbjct 1031 EEEKLELQKLLERVPIPVKESIEEPSAKINVLLQAFISQLKLEGFALMADMVYVTQSAGR 1090
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+MRAIF+I L RGWAQL + L LC I RMW +M PLRQF+ LPEE+++KIEKK+ PF
Sbjct 1091 LMRAIFEIVLNRGWAQLTDKTLNLCKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPF 1150
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+R YDL EIGEL+R+PKMGK +H+ +H FPKLEL+ +QP++RS L VELTITPDFQW
Sbjct 1151 ERLYDLNHNEIGELIRMPKMGKTIHKYVHLFPKLELSVHLQPITRSTLKVELTITPDFQW 1210
Query 181 DQKIHGNGEVFWIL 194
D+K+HG+ E FWIL
Sbjct 1211 DEKVHGSSEAFWIL 1224
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 38/164 (23%), Positives = 78/164 (47%), Gaps = 24/164 (14%)
Query 7 LQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAI 65
L++L ++VP + ++P K N+LLQA++S+++L + +D + A R+++A
Sbjct 1868 LRQLAQKVPHKLNNPKFNDPHVKTNLLLQAHLSRMQLSA-ELQSDTEEILSKAIRLIQAC 1926
Query 66 FDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRK------------I 113
D+ GW A+ A++L + MWS + L+Q E +++ +
Sbjct 1927 VDVLSSNGWLSPALAAMELAQMVTQAMWSKDSYLKQLPHFTSEHIKRCTDKGVESVFDIM 1986
Query 114 EKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELA 157
E +D + LT ++I ++ R + +P +EL+
Sbjct 1987 EMEDEERNALLQLTDSQIADVA----------RFCNRYPNIELS 2020
> hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase-like
Length=1700
Score = 290 bits (742), Expect = 2e-78, Method: Compositional matrix adjust.
Identities = 133/194 (68%), Positives = 163/194 (84%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEKLELQ+L+ERVPIPVK S +EPS+K+NVLLQA+IS+LKLEGFA+MADMVYV QSA R
Sbjct 595 EEEKLELQKLLERVPIPVKESIEEPSAKINVLLQAFISQLKLEGFALMADMVYVTQSAGR 654
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+MRAIF+I L RGWAQL + L LC I RMW +M PLRQF+ LPEE+++KIEKK+ PF
Sbjct 655 LMRAIFEIVLNRGWAQLTDKTLNLCKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPF 714
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+R YDL EIGEL+R+PKMGK +H+ +H FPKLEL+ +QP++RS L VELTITPDFQW
Sbjct 715 ERLYDLNHNEIGELIRMPKMGKTIHKYVHLFPKLELSVHLQPITRSTLKVELTITPDFQW 774
Query 181 DQKIHGNGEVFWIL 194
D+K+HG+ E FWIL
Sbjct 775 DEKVHGSSEAFWIL 788
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/164 (23%), Positives = 78/164 (47%), Gaps = 24/164 (14%)
Query 7 LQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAI 65
L++L ++VP + ++P K N+LLQA++S+++L + +D + A R+++A
Sbjct 1432 LRQLAQKVPHKLNNPKFNDPHVKTNLLLQAHLSRMQLSA-ELQSDTEEILSKAIRLIQAC 1490
Query 66 FDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRK------------I 113
D+ GW A+ A++L + MWS + LR+ P L ++ +
Sbjct 1491 VDVLSSNGWLSPALAAMELAQMVTQAMWSEDSYLRRLPPFPSGLFKRCTDKGVESVFDIM 1550
Query 114 EKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELA 157
E +D + LT ++I ++ R + +P +EL+
Sbjct 1551 EMEDEERNALLQLTDSQIADVA----------RFCNRYPNIELS 1584
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 250 bits (639), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 113/194 (58%), Positives = 157/194 (80%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
++EK+EL +L++RVPIP+K + +EPS+K+NVLLQAYIS+LKLEG ++ +DMVY+ QSA R
Sbjct 1055 QDEKMELAKLLDRVPIPIKETLEEPSAKINVLLQAYISQLKLEGLSLTSDMVYITQSAGR 1114
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
++RA+++I L+RGWAQLA +AL L + RMWS TPLRQF L +++ ++EKKDL +
Sbjct 1115 LVRALYEIVLKRGWAQLAEKALNLSKMVGKRMWSVQTPLRQFHGLSNDILMQLEKKDLVW 1174
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+RYYDL++ E+GEL+R PKMGK LH+ IH FPK+ L+A VQP++R+ L VELT+TPDF W
Sbjct 1175 ERYYDLSAQELGELIRSPKMGKPLHKFIHQFPKVTLSAHVQPITRTVLNVELTVTPDFLW 1234
Query 181 DQKIHGNGEVFWIL 194
D+KIH E FWI+
Sbjct 1235 DEKIHKYVEPFWII 1248
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 64/139 (46%), Gaps = 6/139 (4%)
Query 24 EPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQ 83
+P K N LLQA+ S+ + G M D V SA R+++A+ D+ GW LA+ A++
Sbjct 1913 DPHVKANALLQAHFSRQNIGGNLAM-DQRDVLLSATRLLQAMVDVISSNGWLNLALLAME 1971
Query 84 LCNEIQGRMWSTMTPLRQFKVLPEELMRKI-EKKDLPFDRYYDLTSTEIGELVRVPKMGK 142
+ + MW + L Q ++L ++ E + +DL E E + KM
Sbjct 1972 VSQMVTQGMWERDSMLLQLPHFTKDLAKRCQENPGKNIETVFDLVEMEDEERQELLKMSD 2031
Query 143 L----LHRLIHSFPKLELA 157
+ R + FP ++L
Sbjct 2032 AQLLDIARFCNRFPNIDLT 2050
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 246 bits (629), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 109/194 (56%), Positives = 148/194 (76%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+EE++EL LME+VPIP++G E SSKV VLLQ+YIS+ LEG+A++++M ++ Q+A R
Sbjct 1033 DEERVELSGLMEKVPIPIRGHGQESSSKVAVLLQSYISRFDLEGYALVSEMTFITQNAGR 1092
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
I+RA+++I L W+QLA R C ++ RMWS M PLRQFK LPEEL+ K+E+ D +
Sbjct 1093 ILRALYEIALTNSWSQLAQRLFDFCKMVERRMWSVMLPLRQFKSLPEELILKLERNDFTW 1152
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
DRYYDL+S E+GEL R PK+GK LHRL+H P+LEL FVQPL+R L VE+ I+PDFQW
Sbjct 1153 DRYYDLSSVELGELCRQPKLGKTLHRLVHLVPRLELQVFVQPLTRDVLRVEVGISPDFQW 1212
Query 181 DQKIHGNGEVFWIL 194
DQ++HG+ E FW+
Sbjct 1213 DQRLHGSNERFWLF 1226
Score = 29.6 bits (65), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Query 24 EPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQ 83
EP +KV+ LL A++ + L + +D+ V + ++ A+ D+ GW + A+Q
Sbjct 1889 EPDNKVSELLIAHMQRAMLTN-DLQSDLCLVLERIGALLCALVDVLSSNGWLGPVLLAMQ 1947
Query 84 LCNEI 88
L I
Sbjct 1948 LSQRI 1952
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 246 bits (628), Expect = 4e-65, Method: Compositional matrix adjust.
Identities = 115/194 (59%), Positives = 154/194 (79%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
++EK+EL +L++RVPIPVK + ++PS+K+NVLLQ YISKLKLEG ++ +DMVY+ QSA R
Sbjct 1056 QDEKMELAKLLDRVPIPVKETLEDPSAKINVLLQVYISKLKLEGLSLTSDMVYITQSAGR 1115
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
++RAIF+I L+RGWAQL+ +AL L + RMWS TPL QF +P+E++ K+EK DL +
Sbjct 1116 LLRAIFEIVLKRGWAQLSQKALNLSKMVGKRMWSVQTPLWQFPGIPKEILMKLEKNDLVW 1175
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+RYYDL+S E+GEL+ PKMG+ LH+ IH FPKL+LAA VQP+SRS L VELT+TPDF W
Sbjct 1176 ERYYDLSSQELGELICNPKMGRPLHKYIHQFPKLKLAAHVQPISRSVLQVELTVTPDFHW 1235
Query 181 DQKIHGNGEVFWIL 194
D K + E FWI+
Sbjct 1236 DDKANKYVEPFWII 1249
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 243 bits (621), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 108/194 (55%), Positives = 147/194 (75%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+EEKLELQ++ E PIP+K + DE S+K NVLLQAYIS+LKLEGFA+ ADMV+V QSA R
Sbjct 1024 DEEKLELQKMAEHAPIPIKENLDEASAKTNVLLQAYISQLKLEGFALQADMVFVAQSAGR 1083
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+ RA+F+I L RGWA LA + L LC + R W ++ PL QFK +P E++R I+KK+ F
Sbjct 1084 LFRALFEIVLWRGWAGLAQKVLTLCKMVTQRQWGSLNPLHQFKKIPSEVVRSIDKKNYSF 1143
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
DR YDL ++G+L+++PKMGK L + I FPKLE+ +QP++R+ + +ELTITPDF+W
Sbjct 1144 DRLYDLDQHQLGDLIKMPKMGKPLFKFIRQFPKLEMTTLIQPITRTTMRIELTITPDFKW 1203
Query 181 DQKIHGNGEVFWIL 194
D+K+HG+ E FWI
Sbjct 1204 DEKVHGSAEGFWIF 1217
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 44/167 (26%), Positives = 77/167 (46%), Gaps = 18/167 (10%)
Query 2 EEKLELQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+E + L++L ER+P +K +P KVN+L+ A++S++KL + D + A R
Sbjct 1862 KEDVILRQLAERLPGQLKNQKFTDPHVKVNLLIHAHLSRVKLTA-ELNKDTELIVLRACR 1920
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKK---- 116
+++A D+ GW AI A++L + M+S L+Q L+ + + K
Sbjct 1921 LVQACVDVLSSNGWLSPAIHAMELSQMLTQAMYSNEPYLKQLPHCSAALLERAKAKEVTS 1980
Query 117 -----DLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAA 158
+L D D+ E EL V R + +P +E+A
Sbjct 1981 VFELLELENDDRSDILQMEGAELADVA-------RFCNHYPSIEVAT 2020
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 187 bits (476), Expect = 2e-47, Method: Composition-based stats.
Identities = 88/194 (45%), Positives = 129/194 (66%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+EEK EL +ME++PIPVK S + P +K+N+LLQ Y+S + L G+ + ADMVY+ Q+A R
Sbjct 1429 DEEKTELSIIMEKLPIPVKESINIPYTKINILLQLYLSNIILNGYIINADMVYIHQNALR 1488
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
I R+ F+I L++ L L+ C I+ RMWSTMTPLRQF +L EL+R IEKK++ F
Sbjct 1489 IFRSFFEISLKKNSYNLIKLTLKFCKMIERRMWSTMTPLRQFGLLSTELIRIIEKKNITF 1548
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
Y + E + + K+ K +++L+H FPKLEL A++QP++ L V+L I PDF +
Sbjct 1549 KNYLTMNLNEYITIFKNKKIAKNIYKLVHHFPKLELNAYIQPINHKILKVDLNIAPDFIY 1608
Query 181 DQKIHGNGEVFWIL 194
+ K HG +FW+
Sbjct 1609 NPKYHGYFMLFWVF 1622
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 180 bits (457), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 82/194 (42%), Positives = 127/194 (65%), Gaps = 1/194 (0%)
Query 2 EEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRI 61
EEK EL++L+E+ PIP++ D+P +KVNVLLQ+Y S+LK EGFA+ +D+V++ Q+A R+
Sbjct 1049 EEKRELKQLLEKAPIPIREDIDDPLAKVNVLLQSYFSQLKFEGFALNSDIVFIHQNAGRL 1108
Query 62 MRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFD 121
+RA+F+ICL+RGW L LC +MW T PLRQFK P E+++++E +P+
Sbjct 1109 LRAMFEICLKRGWGHPTRMLLNLCKSATTKMWPTNCPLRQFKTCPVEVIKRLEASTVPWG 1168
Query 122 RYYDL-TSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
Y L T E+G +R K GK ++ L+ FPK+ + QP++RS + + I D+ W
Sbjct 1169 DYLQLETPAEVGRAIRSEKYGKQVYDLLKRFPKMSVTCNAQPITRSVMRFNIEIIADWIW 1228
Query 181 DQKIHGNGEVFWIL 194
D +HG+ E F ++
Sbjct 1229 DMNVHGSLEPFLLM 1242
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 39/157 (24%), Positives = 70/157 (44%), Gaps = 10/157 (6%)
Query 7 LQRLMERVPI--PVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRA 64
L +L +R+P+ P S S KV +LLQAY S+L+L D+ + + ++
Sbjct 1902 LVKLSKRLPLRFPEHTSSGSVSFKVFLLLQAYFSRLELP-VDFQNDLKDILEKVVPLINV 1960
Query 65 IFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYY 124
+ DI G+ A A+ L + +W PLRQ +++ K K++ + Y
Sbjct 1961 VVDILSANGYLN-ATTAMDLAQMLIQGVWDVDNPLRQIPHFNNKILEKC--KEINVETVY 2017
Query 125 DLTSTEIGE----LVRVPKMGKLLHRLIHSFPKLELA 157
D+ + E E L + ++++P +EL
Sbjct 2018 DIMALEDEERDEILTLTDSQLAQVAAFVNNYPNVELT 2054
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 173 bits (438), Expect = 3e-43, Method: Composition-based stats.
Identities = 77/169 (45%), Positives = 119/169 (70%), Gaps = 0/169 (0%)
Query 26 SSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQLC 85
++K+++LLQAYIS+L L+G+A++++M Y+ Q+A RI+ A+F I L+R W+ L+I+ C
Sbjct 1111 NNKISILLQAYISRLDLDGYALVSEMGYITQNAPRILTALFVISLKRCWSSLSIKLFNFC 1170
Query 86 NEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLH 145
++ RMW M PLR FK +P E++ K+EKKD+P+ RYYDL + E+GEL R K+GK L+
Sbjct 1171 KMVESRMWLLMLPLRHFKTIPNEVVTKLEKKDIPWVRYYDLNAVELGELCRNQKLGKSLY 1230
Query 146 RLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQWDQKIHGNGEVFWIL 194
+ +H PK+ L +VQPL+ + + V L I DF WD K H N + F ++
Sbjct 1231 KFVHLVPKVNLQVYVQPLTCNRISVHLVIKRDFVWDYKYHFNYQKFLLV 1279
> mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07Rik,
D630041L21, Helic1, RNAH; activating signal cointegrator
1 complex subunit 3; K01529 [EC:3.6.1.-]
Length=2198
Score = 152 bits (385), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 73/195 (37%), Positives = 118/195 (60%), Gaps = 2/195 (1%)
Query 1 EEEKLELQRLMER-VPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE EL L+ + G + K+N+LLQ YIS+ +++ F++++D YV Q+A
Sbjct 1029 EEEIEELDALLNNFCELSAPGGVENSYGKINILLQTYISRGEMDSFSLISDSAYVAQNAA 1088
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RI+RA+F+I LR+ W + R L L I R+W +PLRQF VLP ++ ++E+K+L
Sbjct 1089 RIVRALFEIALRKRWPTMTYRLLNLSKVIDKRLWGWASPLRQFSVLPPHILTRLEEKNLT 1148
Query 120 FDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQ 179
D+ D+ EIG ++ +G + + +H P + + A +QP++R+ L V L I PDF
Sbjct 1149 VDKLKDMRKDEIGHILHHVNIGLKVKQCVHQIPSVTMEASIQPITRTVLRVSLNIHPDFS 1208
Query 180 WDQKIHGN-GEVFWI 193
W+ ++HG GE +WI
Sbjct 1209 WNDQVHGTVGEPWWI 1223
Score = 37.4 bits (85), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 4/80 (5%)
Query 16 IPVKGSP---DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRR 72
+P++ +P D P +K ++LLQA++S+ L D V A R+ +A+ D+ +
Sbjct 1876 LPIELNPHSFDSPHTKAHLLLQAHLSRAMLPCPDYDTDTKTVLDQALRVCQAMLDVAASQ 1935
Query 73 GWAQLAIRALQLCNE-IQGR 91
GW + L IQGR
Sbjct 1936 GWLVTVLNITHLIQMVIQGR 1955
> hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,
dJ121G13.4; activating signal cointegrator 1 complex subunit
3; K01529 [EC:3.6.1.-]
Length=2202
Score = 147 bits (371), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 71/195 (36%), Positives = 117/195 (60%), Gaps = 2/195 (1%)
Query 1 EEEKLELQRLMER-VPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE EL L+ + G + K+N+LLQ YIS+ +++ F++++D YV Q+A
Sbjct 1028 EEEIEELDTLLSNFCELSTPGGVENSYGKINILLQTYISRGEMDSFSLISDSAYVAQNAA 1087
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RI+RA+F+I LR+ W + R L L I R+W +PLRQF +LP ++ ++E+K L
Sbjct 1088 RIVRALFEIALRKRWPTMTYRLLNLSKVIDKRLWGWASPLRQFSILPPHILTRLEEKKLT 1147
Query 120 FDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQ 179
D+ D+ EIG ++ +G + + +H P + + A +QP++R+ L V L+I DF
Sbjct 1148 VDKLKDMRKDEIGHILHHVNIGLKVKQCVHQIPSVMMEASIQPITRTVLRVTLSIYADFT 1207
Query 180 WDQKIHGN-GEVFWI 193
W+ ++HG GE +WI
Sbjct 1208 WNDQVHGTVGEPWWI 1222
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Query 16 IPVKGSP---DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRR 72
+P++ +P D P +K ++LLQA++S+ L D V A R+ +A+ D+ +
Sbjct 1875 LPIESNPHSFDSPHTKAHLLLQAHLSRAMLPCPDYDTDTKTVLDQALRVCQAMLDVAANQ 1934
Query 73 GWAQLAIRALQLCNEIQ 89
GW + L + N IQ
Sbjct 1935 GW---LVTVLNITNLIQ 1948
> ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6.1.-]
Length=2146
Score = 145 bits (365), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 75/201 (37%), Positives = 115/201 (57%), Gaps = 23/201 (11%)
Query 1 EEEKLELQRLMERV-PIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE+ EL+ L P+ VKG P K+++L+Q YIS+ ++ F++++D Y+ S
Sbjct 1058 EEEQHELETLARSCCPLEVKGGPSNKHGKISILIQLYISRGSIDAFSLVSDASYISASLA 1117
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RIMRA+F+ICLR+GW ++ + L+ C + ++W PLRQF ++DLP
Sbjct 1118 RIMRALFEICLRKGWCEMTLFMLEYCKAVDRQLWPHQHPLRQF------------ERDLP 1165
Query 120 FDR------YYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELT 173
DR Y++ EIG L+R G R + FP ++LAA V P++R+ L V+L
Sbjct 1166 SDRRDDLDHLYEMEEKEIGALIRYNPGG----RHLGYFPSIQLAATVSPITRTVLKVDLL 1221
Query 174 ITPDFQWDQKIHGNGEVFWIL 194
ITP+F W + HG +WIL
Sbjct 1222 ITPNFIWKDRFHGTALRWWIL 1242
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 46/88 (52%), Gaps = 1/88 (1%)
Query 8 QRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIF 66
+ L +RV PV + D+P K N+L QA+ S+L L D+ V + RI++A+
Sbjct 1885 KTLSDRVRYPVDNNHLDDPHVKANLLFQAHFSQLALPISDYNTDLKSVLDQSIRILQAMI 1944
Query 67 DICLRRGWAQLAIRALQLCNEIQGRMWS 94
DIC GW ++ ++L + MWS
Sbjct 1945 DICANSGWLSSSLTCMRLLQMVMQGMWS 1972
> sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, involved
in translation inhibition of non-poly(A) mRNAs; required
for repressing propagation of dsRNA viruses (EC:3.6.1.-);
K01529 [EC:3.6.1.-]
Length=1967
Score = 143 bits (360), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 72/195 (36%), Positives = 114/195 (58%), Gaps = 2/195 (1%)
Query 1 EEEKLELQRLM-ERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE EL+RL E V + D P K NVLLQAYIS+ ++ A+ +D YV Q++
Sbjct 845 EEESKELKRLSDESVECQIGSQLDTPQGKANVLLQAYISQTRIFDSALSSDSNYVAQNSV 904
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RI RA+F I + R W + + L +C I+ R+W+ PL QF LPE ++R+I
Sbjct 905 RICRALFLIGVNRRWGKFSNVMLNICKSIEKRLWAFDHPLCQFD-LPENIIRRIRDTKPS 963
Query 120 FDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQ 179
+ +L + E+GELV K G L++++ FPK+ + A + P++ + + + + + PDF
Sbjct 964 MEHLLELEADELGELVHNKKAGSRLYKILSRFPKINIEAEIFPITTNVMRIHIALGPDFV 1023
Query 180 WDQKIHGNGEVFWIL 194
WD +IHG+ + FW+
Sbjct 1024 WDSRIHGDAQFFWVF 1038
Score = 37.4 bits (85), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 47/97 (48%), Gaps = 5/97 (5%)
Query 24 EPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQ 83
+P K +LLQA++S++ L + D V V + RI++A D+ G+ + ++
Sbjct 1707 DPHVKTFLLLQAHLSRVDLPIADYIQDTVSVLDQSLRILQAYIDVASELGYFHTVLTMIK 1766
Query 84 LCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+ I+ W P+ VLP +R+I KD F
Sbjct 1767 MMQCIKQGYWYEDDPV---SVLPGLQLRRI--KDYTF 1798
> cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-]
Length=1798
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 62/191 (32%), Positives = 108/191 (56%), Gaps = 7/191 (3%)
Query 1 EEEKLELQRLMER-VPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSA 58
EEE +L+ LM + V+G + KVNVLLQ+ IS+ A+M++ +YVQQ+A
Sbjct 658 EEEIGDLEELMSYGCMMNVRGGGLASVAGKVNVLLQSLISRTSTRNSALMSEQLYVQQNA 717
Query 59 NRIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQF---KVLPEELMRKIEK 115
R+ RA+F++ L+ GW+Q A L + ++ +MW LRQF +P + KIE+
Sbjct 718 GRLCRAMFEMVLKNGWSQAANAFLGIAKCVEKQMWMNQCALRQFIQIINIPITWIEKIER 777
Query 116 KDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTIT 175
K +L++ ++G + G L+ + P++++ A +P++ + + VE+T+T
Sbjct 778 KKARESDLLELSAKDLGYMFSCD--GDRLYTYLRYLPRMDVQAKFKPITYTIVQVEVTLT 835
Query 176 PDFQWDQKIHG 186
P F W+ +IHG
Sbjct 836 PTFIWNDQIHG 846
> dre:100332599 mutagen-sensitive 308-like
Length=864
Score = 102 bits (254), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 44/123 (35%), Positives = 74/123 (60%), Gaps = 0/123 (0%)
Query 62 MRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFD 121
MRA+F++ LR+ W + R L LC + R+W PLRQF LP + ++E K+L D
Sbjct 1 MRALFEMALRKRWPAMTYRLLNLCKVMDKRLWGWAHPLRQFNTLPASALARMEDKNLTID 60
Query 122 RYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQWD 181
+ D+ EIG ++ +G + + +H P + L + +QP++R+ L V L+ITPDF+W+
Sbjct 61 KLRDMGKDEIGHMLHHVNIGLKVKQCVHQIPAILLESSIQPITRTVLRVRLSITPDFRWN 120
Query 182 QKI 184
++
Sbjct 121 DQV 123
> tgo:TGME49_049810 activating signal cointegrator 1 complex subunit
3, putative (EC:5.99.1.3)
Length=2539
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 62/213 (29%), Positives = 101/213 (47%), Gaps = 24/213 (11%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQ----Q 56
EE +L R +P+ G D P +KV L+QA +++ ++ F++ AD YVQ
Sbjct 1240 EESELSNLRRSAICRVPIVGDFDAPEAKVQTLVQAALAQAPIKAFSLCADSNYVQAGDAS 1299
Query 57 SANRIMRAIFDICLRRGWAQLAIRALQLCN------EIQGRMW-----STMTPLRQFK-V 104
SA +I+ + + RG + + CN ++Q R + P +Q + V
Sbjct 1300 SAEKILE--WTKAVERGLWPTSHVLMHFCNPNCFDPDVQKRRQPYVPRANEHPGKQNRLV 1357
Query 105 LPEELMRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLS 164
L E ++ ++EK R DL ++EI LV G+ + I P LEL V P++
Sbjct 1358 LREGMVSRLEKHQFALGRLRDLGASEIASLVASKADGQDVALAIRMVPDLELD--VNPIT 1415
Query 165 RSCLVVELTI--TPDFQWDQKIHGNGEVF--WI 193
+ L V + + T +F W HGNGE+F W+
Sbjct 1416 AAILRVSIALRFTEEFLWSAWWHGNGELFHLWV 1448
Score = 29.6 bits (65), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 15 PIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDIC 69
P P+ S + P +K +L QA + +L + D+ +A RI++A+ DIC
Sbjct 2253 PYPIDASTVNSPHTKTFLLFQAQMFQLPVPIADYNTDLKSALDNAMRILQAMLDIC 2308
> cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the second
part of the RNA
Length=1204
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 62/101 (61%), Gaps = 17/101 (16%)
Query 2 EEKLELQRLMERVPIPVKGSPD---------------EPSSKVNVLLQAYISKLK--LEG 44
EEK+EL++L+++VPIP++G + +KVNVLLQ YI+ +
Sbjct 1101 EEKIELEKLVDKVPIPIQGVGSSNVDGNDNIGNMVDLDTFTKVNVLLQLYITGSRWINAK 1160
Query 45 FAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQLC 85
+++D+ ++ QSA RI RAIF++ ++R W+ LA R+L++
Sbjct 1161 LTLLSDLHFIAQSAPRIFRAIFNLAIKRRWSTLARRSLKIA 1201
> xla:100158290 snrnp200, ascc3l1; small nuclear ribonucleoprotein
200kDa (U5); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=457
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 82/156 (52%), Gaps = 8/156 (5%)
Query 7 LQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAI 65
L++L ++VP + ++P K N+LLQA++S+++L + +D + A R+++A
Sbjct 189 LRQLAQKVPHKLTNPKFNDPHVKTNLLLQAHLSRMQLSA-ELQSDTEEILGKAVRLIQAC 247
Query 66 FDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP--FDRY 123
D+ GW A+ A++L + MWS + LRQ E +++ K++ FD
Sbjct 248 VDVLSSNGWLSPALAAMELAQMVTQAMWSKDSYLRQLPHFSSEHIKRCTDKEVESVFD-I 306
Query 124 YDLTSTEIGELVRV--PKMGKLLHRLIHSFPKLELA 157
++ E EL+++ +M + R + +P +EL+
Sbjct 307 MEMEDEERSELLQLSDSQMADVA-RFCNRYPNIELS 341
> tpv:TP01_0582 RNA helicase
Length=1764
Score = 56.2 bits (134), Expect = 7e-08, Method: Composition-based stats.
Identities = 46/208 (22%), Positives = 98/208 (47%), Gaps = 19/208 (9%)
Query 2 EEKLELQRLMERVPIPV---KGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSA 58
EE EL LM P+ + KG + +KV+VL+QAYI+KL ++ +++ D+ ++ Q+
Sbjct 689 EEYDELLDLMNS-PLVIYKPKGGINHIKNKVSVLIQAYIAKLFIKTSSLVTDLNFIVQNI 747
Query 59 NRIMRAIFDICL-----------RRGWAQLAIRALQLCNEIQG--RMWSTMTPLRQFKVL 105
R+ RA F+I + W + R + N + + +TP + +L
Sbjct 748 PRLARAYFEISMCETVCGPPVEHIHDWVLILERQIFNSNVLSNFTSPMNNLTPSKDLGLL 807
Query 106 PEELMRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSR 165
L+ + + L + + + E+ ++VR + + + I P E+ + QP++
Sbjct 808 STNLVDRFNRFKL--EDIINFSYQEVLDIVRSKQDASTISKYIKYIPYPEVKLYNQPITD 865
Query 166 SCLVVELTITPDFQWDQKIHGNGEVFWI 193
+ +++ W ++ +G+ E F++
Sbjct 866 KITKLTVSVEIKNDWSRRWNGSNESFYV 893
> cpv:cgd2_2770 U5 small nuclear ribonucleoprotein 200kDA helicase,
Pre-mRNA splicing helicase BRR2 2 (RNA helicase plus Sec63
domain)
Length=2184
Score = 50.4 bits (119), Expect = 4e-06, Method: Composition-based stats.
Identities = 44/196 (22%), Positives = 91/196 (46%), Gaps = 28/196 (14%)
Query 26 SSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRR--GWAQLAIRALQ 83
SSKV +LL AY ++++ ++ D +Y+ Q+ RI+R IF++ G ++ A R L+
Sbjct 927 SSKVALLLIAYSLRIEITTPTLVMDSIYISQNGARILRFIFELIQLSTFGVSERAQRVLE 986
Query 84 LCNEIQGRMWSTMTPLRQF-------------KVLPEELMRKIEKKDLPF-----DRYYD 125
++ R++ T + LR F + E + K P + D
Sbjct 987 WSKMLEMRIFYTQSVLRHFVYFSSLDKTLNPNETFASERSNRNTKFKGPLKIGSIKKLED 1046
Query 126 LTSTEIGELVRVPKMGKLLH-------RLIHSFPKLELA-AFVQPLSRSCLVVELTITPD 177
S E+ + + + ++ +++ I PK++ A V P++ + + + + P+
Sbjct 1047 YASWEMIKDLAICELKHIVYSDAEKISEYIKYIPKIDFKEALVSPVTLKIVRLGIKLYPN 1106
Query 178 FQWDQKIHGNGEVFWI 193
++W Q+ HG E F++
Sbjct 1107 WKWSQRWHGIREKFYL 1122
> bbo:BBOV_I001530 19.m02171; helicase
Length=1798
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 41/218 (18%), Positives = 90/218 (41%), Gaps = 28/218 (12%)
Query 1 EEEKLELQRLM-ERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
+E EL LM + P +G + ++K+++L++A+I++ ++ ++++DM Y+ Q+
Sbjct 738 NDELEELSDLMRHSIFKPTRG-LNHITTKISLLIEAHINRTYIKSSSLISDMNYIIQNIG 796
Query 60 RIMRAIFDICLRRG------------WAQLAIRAL------------QLCNEIQGRMWST 95
R++ A F++ + WA + R + C
Sbjct 797 RLLLAYFEVSMSETVCAPPIGNLIYKWALMFERQIWDVKLRPLNVLYHFCRPYHAMYDRA 856
Query 96 MTPLRQFKVLPEELMRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLE 155
+ L E ++ +L + DLT +E +LV+ + + P +
Sbjct 857 KMQSSKLPTLSEGTATRLSTYNL--ESLMDLTHSEFSQLVKSRSEASAVESFLGFVPYPQ 914
Query 156 LAAFVQPLSRSCLVVELTITPDFQWDQKIHGNGEVFWI 193
+ +P++ V + IT W + +G E+F+I
Sbjct 915 IIPSSRPITSCITEVNVKITLKNNWSTRWNGKNEIFYI 952
> pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosinetriphosphatase
[EC:3.6.1.3]
Length=2472
Score = 38.5 bits (88), Expect = 0.015, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 35/54 (64%), Gaps = 1/54 (1%)
Query 18 VKGSPDEPSS-KVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICL 70
+K DE + + +L++ Y+ +L++ F+++ ++ Y+ Q+ RI+ A +DICL
Sbjct 1016 IKEEYDESKNMTLRILIEVYLRRLQINNFSIICEINYIVQNIIRILYAYYDICL 1069
> dre:557052 hfm1, si:ch211-83f14.5; HFM1, ATP-dependent DNA helicase
homolog (S. cerevisiae)
Length=1228
Score = 33.5 bits (75), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 31/147 (21%), Positives = 62/147 (42%), Gaps = 5/147 (3%)
Query 16 IPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWA 75
P++G KVN L+QA + + ++ F ++ D + ++A R+ R + + +
Sbjct 736 FPIEGKIKSNDMKVNCLIQAQLGCIPIQEFGLIQDTGKIFRNAVRVSRYLTEFLCHHSKS 795
Query 76 QLA--IRALQLCNEIQGRMWSTMTPL-RQFKVLPEELMRKIEKKDLPFDRYYDLTSTEIG 132
A + AL L + ++W + +Q + + L + L + TS
Sbjct 796 NFAAQLNALILAKCFRAKLWENSPYISKQLERIGLSLATALVNAGLTTFSKIEQTSPREL 855
Query 133 ELV--RVPKMGKLLHRLIHSFPKLELA 157
EL+ R P G + I P+ E++
Sbjct 856 ELIVNRHPPFGNQIKEAISKLPRCEVS 882
> mmu:446101 Xrra1, AI449753; X-ray radiation resistance associated
1
Length=786
Score = 33.5 bits (75), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 52/107 (48%), Gaps = 9/107 (8%)
Query 45 FAMMADMVYVQQ---SANRIMRAIF--DICLRRGWAQLAIRALQLCNEIQGRMWSTMTPL 99
FA +A + +++ N+I+R + I LR G E+Q +MW TP
Sbjct 251 FASLAGLRRLKKLSLDQNKIVRIPYLQQIQLRDGSGDWVTEGPNPQKELQPQMWIFETPD 310
Query 100 RQ--FKVLPEELMRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 144
Q + VLP + + +++ ++ F Y +++E ++ +P M ++L
Sbjct 311 EQPNYTVLP--MKKDVDRTEVVFSSYPGFSTSETAKVCSLPPMFEIL 355
> bbo:BBOV_IV010770 23.m05766; hypothetical protein
Length=1104
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 83 QLCNEIQGRMWSTMTPLRQFKV--LPEELMRKIEK----KDLPFDRYYDLTSTEIGELVR 136
Q+C+EI +MWS++ ++ +V L E+ +E+ KD RY T ++ +L
Sbjct 616 QICDEIDLQMWSSIIGQKEKRVQKLRSEVETTVERLSSFKDTIESRYSSYTRKDLNDLKH 675
Query 137 VPKMGK 142
+P K
Sbjct 676 IPGFAK 681
> hsa:164045 HFM1, FLJ36760, FLJ39011, MER3, MGC163397, RP11-539G11.1,
SEC63D1, Si-11, Si-11-6, helicase; HFM1, ATP-dependent
DNA helicase homolog (S. cerevisiae) (EC:3.6.4.12)
Length=1435
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 37/167 (22%), Positives = 74/167 (44%), Gaps = 7/167 (4%)
Query 14 VPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFD-ICLRR 72
+ P++G KVN L+QA + + ++ FA+ D + + +RI R + D + +
Sbjct 844 IRFPMEGRIKTREMKVNCLIQAQLGCIPIQDFALTQDTAKIFRHGSRITRWLSDFVAAQE 903
Query 73 GWAQLAIRALQLCNEIQGRMW-STMTPLRQFKVLPEELMRKIEKKDLPFDRYYDLTSTEI 131
+ + +L L + ++W +++ +Q + + L I L + + T
Sbjct 904 KKFAVLLNSLILAKCFRCKLWENSLHVSKQLEKIGITLSNAIVNAGLTSFKKIEETDARE 963
Query 132 GELV--RVPKMGKLLHRLIHSFPKLELAAFVQPLSR-SCLVVELTIT 175
EL+ R P G + + PK EL V+ ++R S E+ +T
Sbjct 964 LELILNRHPPFGTQIKETVMYLPKYELK--VEQITRYSDTTAEILVT 1008
> mmu:330149 Hfm1, A330009G12Rik, Gm1046, Sec63d1; HFM1, ATP-dependent
DNA helicase homolog (S. cerevisiae)
Length=1434
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 40/183 (21%), Positives = 80/183 (43%), Gaps = 24/183 (13%)
Query 28 KVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFD-ICLRRGWAQLAIRALQLCN 86
KVN L+QA + + ++ FA+ D V + ++ +RI R + D + + + + ++ L
Sbjct 856 KVNCLIQAQLGCIPIQDFALTQDTVKIFRNGSRIARWLSDFVAAQEKKFAVLLNSVILTK 915
Query 87 EIQGRMWSTMTPL-RQFKVLPEELMRKIEKKDL-PFDRYYDLTSTEIGELV--RVPKMGK 142
+ ++W + +Q + L + L F + + + E+ EL+ R P G
Sbjct 916 CFKCKLWENSKHVSKQLDKIGISLSNTMVNAGLTSFKKIEEANAREL-ELILNRHPPFGT 974
Query 143 LLHRLIHSFPKLELAAFVQPLSR-SCLVVELTIT---------------PDFQWDQKIHG 186
+ + PK EL V+ ++R S + E+ +T PDF + I G
Sbjct 975 QIKEAVAHLPKYELE--VEQIARYSDIKAEILVTIILRNFEQLQTKRTAPDFHYATLIIG 1032
Query 187 NGE 189
+ +
Sbjct 1033 DAD 1035
Lambda K H
0.324 0.139 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5608917492
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40