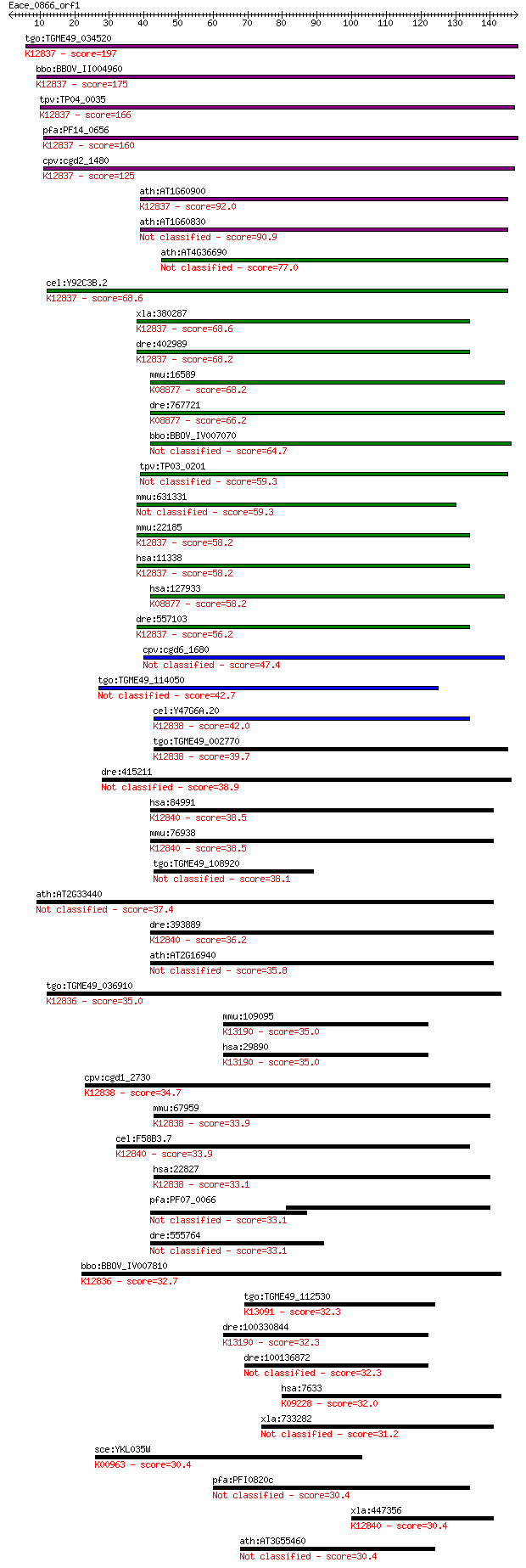
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0866_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_034520 U2 snRNP auxiliary factor or splicing factor... 197 9e-51
bbo:BBOV_II004960 18.m06413; RNA recognition motif (RRM)-conta... 175 4e-44
tpv:TP04_0035 U2 small nuclear ribonucleoprotein, auxiliary fa... 166 2e-41
pfa:PF14_0656 U2 snRNP auxiliary factor, putative; K12837 spli... 160 2e-39
cpv:cgd2_1480 splicing factor U2AF U2 SnRNP auxiliary factor l... 125 7e-29
ath:AT1G60900 U2 snRNP auxiliary factor large subunit, putativ... 92.0 5e-19
ath:AT1G60830 U2 snRNP auxiliary factor large subunit, putative 90.9 1e-18
ath:AT4G36690 ATU2AF65A; RNA binding / nucleic acid binding / ... 77.0 2e-14
cel:Y92C3B.2 uaf-1; U2AF splicing factor family member (uaf-1)... 68.6 7e-12
xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxil... 68.6 7e-12
dre:402989 u2af2b, MGC77804, wu:fb73g02, zgc:77804; U2 small n... 68.2 9e-12
mmu:16589 Uhmk1, 4732477C12Rik, 4930500M09Rik, AA673513, AI449... 68.2 1e-11
dre:767721 uhmk1, MGC153241, zgc:153241; U2AF homology motif (... 66.2 3e-11
bbo:BBOV_IV007070 23.m05938; hypothetical protein 64.7 9e-11
tpv:TP03_0201 hypothetical protein 59.3 4e-09
mmu:631331 Gm7061, EG631331; predicted gene 7061 59.3
mmu:22185 U2af2, 65kDa, MGC118033; U2 small nuclear ribonucleo... 58.2 9e-09
hsa:11338 U2AF2, U2AF65; U2 small nuclear RNA auxiliary factor... 58.2 9e-09
hsa:127933 UHMK1, DKFZp434C1613, FLJ23015, KIS, KIST; U2AF hom... 58.2 9e-09
dre:557103 u2af2a, MGC110115, zgc:110115; U2 small nuclear RNA... 56.2 4e-08
cpv:cgd6_1680 splicing factor U2AF U2 snRNP auxiliary factor l... 47.4 2e-05
tgo:TGME49_114050 splicing factor U2AF 65 kDa subunit, putative 42.7 4e-04
cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing f... 42.0 7e-04
tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(... 39.7 0.003
dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.... 38.9 0.005
hsa:84991 RBM17, DKFZp686F13131, MGC14439, SPF45; RNA binding ... 38.5 0.008
mmu:76938 Rbm17, 2700027J02Rik; RNA binding motif protein 17; ... 38.5 0.008
tgo:TGME49_108920 U2 small nuclear ribonucleoprotein auxiliary... 38.1 0.010
ath:AT2G33440 splicing factor family protein 37.4 0.016
dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif prote... 36.2 0.039
ath:AT2G16940 RNA recognition motif (RRM)-containing protein 35.8 0.046
tgo:TGME49_036910 U2 snRNP auxiliary factor small subunit, put... 35.0 0.082
mmu:109095 Rbm15b, 1810017N16Rik; RNA binding motif protein 15... 35.0 0.092
hsa:29890 RBM15B, HUMAGCGB, OTT3; RNA binding motif protein 15... 35.0 0.093
cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA bind... 34.7 0.11
mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U... 33.9 0.19
cel:F58B3.7 hypothetical protein; K12840 splicing factor 45 33.9
hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding... 33.1 0.29
pfa:PF07_0066 conserved Plasmodium protein, unknown function 33.1 0.32
dre:555764 gon4l, fb99g07, udu, wu:fb99g07; gon-4-like (C.eleg... 33.1 0.34
bbo:BBOV_IV007810 23.m06352; U2 splicing factor subunit; K1283... 32.7 0.43
tgo:TGME49_112530 splicing factor protein, putative ; K13091 R... 32.3 0.48
dre:100330844 mCG19533-like; K13190 RNA-binding protein 15 32.3
dre:100136872 rbm15b, zgc:175165; RNA binding motif protein 15B 32.3 0.59
hsa:7633 ZNF79, pT7; zinc finger protein 79; K09228 KRAB domai... 32.0 0.78
xla:733282 rbm17; RNA binding motif protein 17 31.2
sce:YKL035W UGP1; UDP-glucose pyrophosphorylase (UGPase), cata... 30.4 1.9
pfa:PFI0820c RNA binding protein, putative 30.4 1.9
xla:447356 MGC130690; MGC84102 protein; K12840 splicing factor 45 30.4
ath:AT3G55460 SCL30; SCL30; RNA binding / nucleic acid binding... 30.4 2.1
> tgo:TGME49_034520 U2 snRNP auxiliary factor or splicing factor,
putative ; K12837 splicing factor U2AF 65 kDa subunit
Length=553
Score = 197 bits (501), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 91/142 (64%), Positives = 114/142 (80%), Gaps = 0/142 (0%)
Query 6 QHTTEVTALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEA 65
T VT+LP+S + +LSDP+VA+QV+A R IGE+PS++VQLLN VY ED++ +EA
Sbjct 412 NRTMAVTSLPNSMTQKLLSDPLVAVQVQAARKIGERPSKVVQLLNCVYQEDLIDPKEYEA 471
Query 66 AVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQ 125
DI+ EAEK+G LE+V++P+PNEDLSY+ GVGKVFL Y DVT+AR+AQLMLNGRRFD
Sbjct 472 ICDDIKQEAEKHGALEEVLVPKPNEDLSYREGVGKVFLRYSDVTAARKAQLMLNGRRFDS 531
Query 126 TRVVCAAFFPEERFKEGHYTLT 147
RVVCAAFFPEE+F G YTLT
Sbjct 532 NRVVCAAFFPEEKFAAGRYTLT 553
> bbo:BBOV_II004960 18.m06413; RNA recognition motif (RRM)-containing
protein; K12837 splicing factor U2AF 65 kDa subunit
Length=383
Score = 175 bits (444), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 80/138 (57%), Positives = 105/138 (76%), Gaps = 0/138 (0%)
Query 9 TEVTALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVK 68
+ T LP+S + ++LS+P++ +Q+++GR IG KPSRIVQL+N V+ ED++ D +
Sbjct 245 CKATNLPNSVTQSILSNPLLGLQMQSGRRIGSKPSRIVQLINIVFHEDLIQDKRYHEVKD 304
Query 69 DIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRV 128
I EA+KYG LED+VIPRPN+DLSYK GVGKVFL +GD S+RRAQ MLNGR FD R+
Sbjct 305 AIMEEAKKYGHLEDIVIPRPNDDLSYKEGVGKVFLKFGDEISSRRAQYMLNGRVFDGNRI 364
Query 129 VCAAFFPEERFKEGHYTL 146
VCAAFFP +RF +G YTL
Sbjct 365 VCAAFFPLDRFLKGKYTL 382
> tpv:TP04_0035 U2 small nuclear ribonucleoprotein, auxiliary
factor, large subunit; K12837 splicing factor U2AF 65 kDa subunit
Length=380
Score = 166 bits (421), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 70/137 (51%), Positives = 108/137 (78%), Gaps = 0/137 (0%)
Query 10 EVTALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVKD 69
+ + LP+S + ++LS+P++ +Q++ GR IG PS+++QLLN V+ ED+++D ++ V+
Sbjct 243 KASNLPNSITQSILSNPLLGLQLQNGRRIGSNPSKVIQLLNMVFHEDLISDYNYNEIVRL 302
Query 70 IRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVV 129
++ EA+KYGPL++VVIPRP++DL++K GVGKVF+ Y D+ SAR+AQ M NGR FD+ R+V
Sbjct 303 VKEEAQKYGPLQEVVIPRPDKDLTFKEGVGKVFIRYEDLLSARKAQYMFNGRVFDKNRIV 362
Query 130 CAAFFPEERFKEGHYTL 146
C+AFFPE+ F G YTL
Sbjct 363 CSAFFPEDLFITGKYTL 379
> pfa:PF14_0656 U2 snRNP auxiliary factor, putative; K12837 splicing
factor U2AF 65 kDa subunit
Length=833
Score = 160 bits (404), Expect = 2e-39, Method: Composition-based stats.
Identities = 74/137 (54%), Positives = 104/137 (75%), Gaps = 1/137 (0%)
Query 11 VTALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDI 70
VT LPSS S +LS+ ++ +QV+A R IGEK S++VQL N V+ ED++ D+ +E +K++
Sbjct 698 VTLLPSSISQKILSNSIIGLQVQASRKIGEKSSKVVQLTNAVFQEDLIVDSQYEEILKEV 757
Query 71 RSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVC 130
+ EAEKYG L+++VIP+PN+DLSY GVGK+FL Y D +AR+AQ M NGR F++ RVVC
Sbjct 758 KEEAEKYGTLQNIVIPKPNKDLSYTEGVGKIFLHYADEATARKAQYMFNGRLFEK-RVVC 816
Query 131 AAFFPEERFKEGHYTLT 147
AAF+ EE F +G Y L+
Sbjct 817 AAFYSEEHFLKGKYVLS 833
> cpv:cgd2_1480 splicing factor U2AF U2 SnRNP auxiliary factor
large subunit, RRM domain ; K12837 splicing factor U2AF 65
kDa subunit
Length=438
Score = 125 bits (313), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 53/138 (38%), Positives = 89/138 (64%), Gaps = 2/138 (1%)
Query 11 VTALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDI 70
+T +P+S +Y + S+PV+ + ++ + +GE PS+I+QLLN PE+++ + + + + +
Sbjct 290 ITEIPTSMTYNIFSNPVLGLMMKYSKQVGETPSQIIQLLNIFLPEELVDNEIYNSTLDSV 349
Query 71 RSEAEKYGPLEDVVIPRPN--EDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRV 128
RSEAE YG + ++ PRP E+ G GKVF+ + D+T+ARRAQ NGR FD +
Sbjct 350 RSEAEVYGTILEIFCPRPKVIEEFHSCSGAGKVFIYFSDITAARRAQYQFNGRVFDNIKT 409
Query 129 VCAAFFPEERFKEGHYTL 146
V A FFP E++ + Y++
Sbjct 410 VSATFFPLEKYLKHEYSV 427
> ath:AT1G60900 U2 snRNP auxiliary factor large subunit, putative;
K12837 splicing factor U2AF 65 kDa subunit
Length=589
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 67/106 (63%), Gaps = 1/106 (0%)
Query 39 GEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGV 98
G P++IV L V +D+ D + ++D+R E K+G L +VVIPRPN D PGV
Sbjct 483 GGTPTKIVCLTQVVTADDLRDDEEYAEIMEDMRQEGGKFGNLVNVVIPRPNPDHDPTPGV 542
Query 99 GKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHY 144
GKVFL Y DV + +A+ +NGR+F +VV A ++PE+++ +G Y
Sbjct 543 GKVFLEYADVDGSSKARSGMNGRKFGGNQVV-AVYYPEDKYAQGDY 587
> ath:AT1G60830 U2 snRNP auxiliary factor large subunit, putative
Length=111
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 67/106 (63%), Gaps = 1/106 (0%)
Query 39 GEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGV 98
G P++IV L V +D+ DA + ++D+ E K+G L +VVIPRPN D PGV
Sbjct 5 GGTPTKIVCLTQVVTADDLRDDAEYADIMEDMSQEGGKFGNLVNVVIPRPNPDHDPTPGV 64
Query 99 GKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHY 144
GKVFL Y DV + +A+ +NGR+F +VV A ++PE+++ +G Y
Sbjct 65 GKVFLEYADVDGSSKARSGMNGRKFGGNQVV-AVYYPEDKYAQGDY 109
> ath:AT4G36690 ATU2AF65A; RNA binding / nucleic acid binding
/ nucleotide binding
Length=573
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 61/100 (61%), Gaps = 1/100 (1%)
Query 45 IVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLV 104
+V L V +++ D + ++D+R E K+G L +VVIPRP+ + G+GKVFL
Sbjct 473 VVCLTQVVTEDELRDDEEYGDIMEDMRQEGGKFGALTNVVIPRPSPNGEPVAGLGKVFLK 532
Query 105 YGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHY 144
Y D + RA+ +NGR+F VV A ++PE++F++G Y
Sbjct 533 YADTDGSTRARFGMNGRKFGGNEVV-AVYYPEDKFEQGDY 571
> cel:Y92C3B.2 uaf-1; U2AF splicing factor family member (uaf-1);
K12837 splicing factor U2AF 65 kDa subunit
Length=474
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 69/133 (51%), Gaps = 11/133 (8%)
Query 12 TALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIR 71
T LP+S S + I + G + + I+ L+N V +++ D +E ++D+R
Sbjct 353 TNLPNSAS------AIAGIDLSQG---AGRATEILCLMNMVTEDELKADDEYEEILEDVR 403
Query 72 SEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCA 131
E KYG + + IPRP ED PGVGKVF+ + + +RAQ L GR+F R V
Sbjct 404 DECSKYGIVRSLEIPRPYEDHPV-PGVGKVFVEFASTSDCQRAQAALTGRKF-ANRTVVT 461
Query 132 AFFPEERFKEGHY 144
+++ +++ +
Sbjct 462 SYYDVDKYHNRQF 474
> xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxiliary
factor 2; K12837 splicing factor U2AF 65 kDa subunit
Length=456
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 57/96 (59%), Gaps = 2/96 (2%)
Query 38 IGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPG 97
+G P+ ++ L+N V PE+++ D +E V+D+R E KYG ++ + IPRP + + PG
Sbjct 351 MGGHPTEVLCLMNMVVPEELIDDDEYEEIVEDVRDECGKYGAVKSIEIPRPVDGVEV-PG 409
Query 98 VGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAF 133
GK+F+ + V ++A L GR+F RVV +
Sbjct 410 CGKIFVEFTSVFDCQKAMQGLTGRKF-ANRVVVTKY 444
> dre:402989 u2af2b, MGC77804, wu:fb73g02, zgc:77804; U2 small
nuclear RNA auxiliary factor 2b; K12837 splicing factor U2AF
65 kDa subunit
Length=475
Score = 68.2 bits (165), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 58/96 (60%), Gaps = 2/96 (2%)
Query 38 IGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPG 97
+G P+ ++ L+N V PE+++ D +E V+D++ E KYG ++ + IPRP + L PG
Sbjct 370 MGGIPTEVLCLMNMVAPEELIDDEEYEEIVEDVKEECSKYGQVKSIEIPRPVDGLDI-PG 428
Query 98 VGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAF 133
GK+F+ + V +++A L GR+F RVV +
Sbjct 429 TGKIFVEFTSVYDSQKAMQGLTGRKF-ANRVVVTKY 463
> mmu:16589 Uhmk1, 4732477C12Rik, 4930500M09Rik, AA673513, AI449218,
AU021979, KIS, Kist; U2AF homology motif (UHM) kinase
1 (EC:2.7.11.1); K08877 U2AF homology motif (UHM) kinase 1
[EC:2.7.11.1]
Length=419
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 61/102 (59%), Gaps = 7/102 (6%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKV 101
P+ +++LLN + + + + +E V+D++ E +KYGP+ +++P+ N PG G+V
Sbjct 319 PTPVLRLLNVLDDDYLENEDEYEDVVEDVKEECQKYGPVVSLLVPKEN------PGRGQV 372
Query 102 FLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGH 143
F+ Y + ++ AQ +L GR FD VV A F+P +K G+
Sbjct 373 FVEYANAGDSKAAQKLLTGRMFDGKFVV-ATFYPLSAYKRGY 413
> dre:767721 uhmk1, MGC153241, zgc:153241; U2AF homology motif
(UHM) kinase 1 (EC:2.7.11.1); K08877 U2AF homology motif (UHM)
kinase 1 [EC:2.7.11.1]
Length=410
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 59/102 (57%), Gaps = 7/102 (6%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKV 101
P+ +++LLN + + + +E ++D++ E +KYG + ++IP+ N PG G+V
Sbjct 310 PTPVLRLLNVIDDSHLYNEDEYEDIIEDMKEECQKYGTVVSLLIPKEN------PGKGQV 363
Query 102 FLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGH 143
F+ Y + ++ AQ +L GR FD + V A F+P +K G+
Sbjct 364 FVEYANAGDSKEAQRLLTGRTFD-GKFVVATFYPLGAYKRGY 404
> bbo:BBOV_IV007070 23.m05938; hypothetical protein
Length=400
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 61/111 (54%), Gaps = 8/111 (7%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYK------ 95
P+R++ L N V ED+ DA + + D+R E E+YGP+ V +PR + L+
Sbjct 254 PTRVLLLANLVSKEDLEDDAEYYDIIDDVRCECEEYGPVVRVEMPRVPKGLTLDEIRNMD 313
Query 96 -PGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHYT 145
VG F+++ ++ A +A+ +L+GR+F R+V FF E F G ++
Sbjct 314 FSAVGCAFVLFSNIEGASKARKVLDGRKFGH-RIVECHFFSELLFHVGEFS 363
> tpv:TP03_0201 hypothetical protein
Length=509
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 61/113 (53%), Gaps = 8/113 (7%)
Query 39 GEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLS----- 93
E P+R++ L N V +++ D + + D+R E E YG + V +PR + L+
Sbjct 355 AEIPTRVLLLSNLVSKDELEDDEEYVDIIDDVRCECELYGVVLRVELPRVPKGLTEEEMK 414
Query 94 -YKP-GVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHY 144
+ P VG F+++ V SA +A+ +L+GR+F Q R V A FF E F G +
Sbjct 415 AFDPTSVGSAFVLFSTVESASKARKVLDGRKFGQ-RTVHAHFFSELYFLTGKF 466
> mmu:631331 Gm7061, EG631331; predicted gene 7061
Length=730
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 54/92 (58%), Gaps = 1/92 (1%)
Query 38 IGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPG 97
+G P+ ++ L+N V P++++ D +E V D+R E KYG ++ + IPRP + + PG
Sbjct 625 MGGHPTTVLCLMNMVLPKELLDDEEYEEIVDDVRDECNKYGLVKSIEIPRPMDGVEV-PG 683
Query 98 VGKVFLVYGDVTSARRAQLMLNGRRFDQTRVV 129
GK+F+ + V ++A L GR+F VV
Sbjct 684 CGKIFVEFTSVIDCQKAMQGLTGRKFANRVVV 715
> mmu:22185 U2af2, 65kDa, MGC118033; U2 small nuclear ribonucleoprotein
auxiliary factor (U2AF) 2; K12837 splicing factor
U2AF 65 kDa subunit
Length=475
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 57/96 (59%), Gaps = 2/96 (2%)
Query 38 IGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPG 97
+G P+ ++ L+N V PE+++ D +E V+D+R E KYG ++ + IPRP + + PG
Sbjct 370 MGGHPTEVLCLMNMVLPEELLDDEEYEEIVEDVRDECSKYGLVKSIEIPRPVDGVEV-PG 428
Query 98 VGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAF 133
GK+F+ + V ++A L GR+F RVV +
Sbjct 429 CGKIFVEFTSVFDCQKAMQGLTGRKF-ANRVVVTKY 463
> hsa:11338 U2AF2, U2AF65; U2 small nuclear RNA auxiliary factor
2; K12837 splicing factor U2AF 65 kDa subunit
Length=471
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 57/96 (59%), Gaps = 2/96 (2%)
Query 38 IGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPG 97
+G P+ ++ L+N V PE+++ D +E V+D+R E KYG ++ + IPRP + + PG
Sbjct 366 MGGHPTEVLCLMNMVLPEELLDDEEYEEIVEDVRDECSKYGLVKSIEIPRPVDGVEV-PG 424
Query 98 VGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAF 133
GK+F+ + V ++A L GR+F RVV +
Sbjct 425 CGKIFVEFTSVFDCQKAMQGLTGRKF-ANRVVVTKY 459
> hsa:127933 UHMK1, DKFZp434C1613, FLJ23015, KIS, KIST; U2AF homology
motif (UHM) kinase 1 (EC:2.7.11.1); K08877 U2AF homology
motif (UHM) kinase 1 [EC:2.7.11.1]
Length=345
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 61/102 (59%), Gaps = 7/102 (6%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKV 101
P+ +++LLN + + + + +E V+D++ E +KYGP+ +++P+ N PG G+V
Sbjct 245 PTPVLRLLNVLDDDYLENEEEYEDVVEDVKEECQKYGPVVSLLVPKEN------PGRGQV 298
Query 102 FLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGH 143
F+ Y + ++ AQ +L GR FD VV A F+P +K G+
Sbjct 299 FVEYANAGDSKAAQKLLTGRMFDGKFVV-ATFYPLSAYKRGY 339
> dre:557103 u2af2a, MGC110115, zgc:110115; U2 small nuclear RNA
auxiliary factor 2a; K12837 splicing factor U2AF 65 kDa subunit
Length=465
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 57/96 (59%), Gaps = 2/96 (2%)
Query 38 IGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPG 97
+G P+ ++ L+N V PE+++ D +E V+D+R E KYG ++ + IPRP + L PG
Sbjct 360 MGGIPTEVLCLMNMVAPEELLDDEEYEEIVEDVRDECSKYGQVKSIEIPRPVDGLDI-PG 418
Query 98 VGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAF 133
GK+F+ + V +++A L GR+ RVV +
Sbjct 419 TGKIFVEFMSVFDSQKAMQGLTGRK-SANRVVVTKY 453
> cpv:cgd6_1680 splicing factor U2AF U2 snRNP auxiliary factor
large subunit; 3 RRM domains
Length=492
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 31/109 (28%), Positives = 56/109 (51%), Gaps = 6/109 (5%)
Query 40 EKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYK---- 95
+KP R + L N + E+++ +++ + ++I + KYG + IP P LS K
Sbjct 357 QKPCRCILLSNILTVEELLIPSTYSSIHEEIHEKCLKYGEIYKTTIPIPERALSNKDQFN 416
Query 96 -PGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGH 143
P G+ F+ + +V SA +A+L L RF R + +++ E F G+
Sbjct 417 DPYFGRAFIFFYNVESAIKAKLDLFKMRF-LGRNMKISYYCEHEFLHGN 464
> tgo:TGME49_114050 splicing factor U2AF 65 kDa subunit, putative
Length=414
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 47/99 (47%), Gaps = 5/99 (5%)
Query 27 VVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIP 86
V+ Q++A G+ PSR+V++ P + D ++ + DI +E KYG +I
Sbjct 170 VIQSQIQADLLSGQ-PSRVVRI---ACPSKIERDEEYDEILSDILAECNKYGHALAALII 225
Query 87 RPN-EDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFD 124
RP E L VG V+L Y A L +GR +D
Sbjct 226 RPELEQLLPSVTVGDVYLEYASCIQADHIILTFSGRMYD 264
> cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing
family member (rnp-6); K12838 poly(U)-binding-splicing factor
PUF60
Length=749
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 42/91 (46%), Gaps = 10/91 (10%)
Query 43 SRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVF 102
S ++ L N V P+D+ E +IR E KYG + DVVI G+ K+F
Sbjct 657 SNVIVLRNMVTPQDI-----DEFLEGEIREECGKYGNVIDVVIAN-----FASSGLVKIF 706
Query 103 LVYGDVTSARRAQLMLNGRRFDQTRVVCAAF 133
+ Y D RA+ L+GR F V A+
Sbjct 707 VKYSDSMQVDRAKAALDGRFFGGNTVKAEAY 737
> tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(U)-binding-splicing
factor PUF60
Length=532
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 51/103 (49%), Gaps = 15/103 (14%)
Query 43 SRIVQLLNTVYPEDVMTDASHEAAVKD-IRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKV 101
S +V L N V P +V + +KD +R E K+G ++ V + E + ++
Sbjct 442 SPVVLLSNMVTPSEV------DGELKDEVREECSKFGSIKRVEVHTLKETV-------RI 488
Query 102 FLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHY 144
F+ + D++ AR A L+GR F R + A + +E F +G Y
Sbjct 489 FVEFSDLSGAREAIPSLHGRWFG-GRQIIANTYDQELFHQGEY 530
> dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.2,
si:zc12p8.2, wu:fb33e11, wu:fe37c05, zgc:86806; poly-U
binding splicing factor b
Length=502
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 55/121 (45%), Gaps = 10/121 (8%)
Query 28 VAIQVRAGRTIG-EKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVI- 85
+++ AGR + S ++ L N V PED+ D E + E KYG + V+I
Sbjct 388 LSVSASAGRQAAIQSKSTVMVLRNMVGPEDIDDDLEGE-----VMEECGKYGAVNRVIIY 442
Query 86 -PRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFKEGHY 144
R E+ + V K+F+ + D +A LN R F +VV A + ++RF
Sbjct 443 QERQGEEDDAEIIV-KIFVEFSDAGEMNKAIQALNNRWFAGRKVV-AELYDQDRFNSSDL 500
Query 145 T 145
T
Sbjct 501 T 501
> hsa:84991 RBM17, DKFZp686F13131, MGC14439, SPF45; RNA binding
motif protein 17; K12840 splicing factor 45
Length=401
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 51/102 (50%), Gaps = 14/102 (13%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVI---PRPNEDLSYKPGV 98
P+++V L N V +V D E + E EKYG + VI P +D + +
Sbjct 303 PTKVVLLRNMVGAGEVDEDLEVET-----KEECEKYGKVGKCVIFEIPGAPDDEAVR--- 354
Query 99 GKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFK 140
+FL + V SA +A + LNGR F RVV A F+ ++F+
Sbjct 355 --IFLEFERVESAIKAVVDLNGRYFG-GRVVKACFYNLDKFR 393
> mmu:76938 Rbm17, 2700027J02Rik; RNA binding motif protein 17;
K12840 splicing factor 45
Length=405
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 51/102 (50%), Gaps = 14/102 (13%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVI---PRPNEDLSYKPGV 98
P+++V L N V +V D E + E EKYG + VI P +D + +
Sbjct 307 PTKVVLLRNMVGAGEVDEDLEVE-----TKEECEKYGKVGKCVIFEIPGAPDDEAVR--- 358
Query 99 GKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFK 140
+FL + V SA +A + LNGR F RVV A F+ ++F+
Sbjct 359 --IFLEFERVESAIKAVVDLNGRYFG-GRVVKACFYNLDKFR 397
> tgo:TGME49_108920 U2 small nuclear ribonucleoprotein auxiliary
factor U2AF
Length=704
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 43 SRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKY-GPLEDVVIPRP 88
+R++ L N V ED++ D +E V+DIR E E+ GP+ V IPRP
Sbjct 424 TRVLLLSNIVEVEDLLDDKEYEDIVEDIRLECEECGGPVLSVNIPRP 470
> ath:AT2G33440 splicing factor family protein
Length=247
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 64/135 (47%), Gaps = 6/135 (4%)
Query 9 TEVTALPSSTSYAVLSDP-VVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAA- 66
T V A P ++S AV +P I A +G KP I++L N V PED+ + + E
Sbjct 110 TAVCAFPDASSVAVNENPPFYGIPSHAKPLLG-KPKNILKLKNVVDPEDLTSFSEQEVKE 168
Query 67 -VKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQ 125
++D+R E ++ D + ED G +F+ Y + A L+GR +D
Sbjct 169 ILEDVRLECARWDA-GDKIEEEQEEDPEDVFETGCIFIEYRRPEATCDAAHSLHGRLYDN 227
Query 126 TRVVCAAFFPEERFK 140
R+V A + +E ++
Sbjct 228 -RIVKAEYVSKELYQ 241
> dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif protein
17; K12840 splicing factor 45
Length=418
Score = 36.2 bits (82), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 51/99 (51%), Gaps = 8/99 (8%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKV 101
P+++V L N V +V D E + E EKYG + VI + ++ V ++
Sbjct 320 PTKVVLLRNMVGRGEVDEDLEAE-----TKEECEKYGKVVRCVIFEIS-GVTDDEAV-RI 372
Query 102 FLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFK 140
FL + V SA +A + LNGR F RVV A F+ +RF+
Sbjct 373 FLEFERVESAIKAVVDLNGRYFG-GRVVKACFYNLDRFR 410
> ath:AT2G16940 RNA recognition motif (RRM)-containing protein
Length=561
Score = 35.8 bits (81), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 9/99 (9%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKV 101
PS + L N P D E +D++ E K+G L + + K VG V
Sbjct 463 PSECLLLKNMFDPSTETEDDFDEDIKEDVKEECSKFGKLNHIFVD--------KNSVGFV 514
Query 102 FLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFK 140
+L + + +A AQ L+GR F +++ A + E ++
Sbjct 515 YLRFENAQAAIGAQRALHGRWF-AGKMITATYMTTEAYE 552
> tgo:TGME49_036910 U2 snRNP auxiliary factor small subunit, putative
; K12836 splicing factor U2AF 35 kDa subunit
Length=254
Score = 35.0 bits (79), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 59/132 (44%), Gaps = 20/132 (15%)
Query 12 TALPSSTSYAVLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASH-EAAVKDI 70
T+ P+ + +P VA+ + G+ + + +LL+ A H EA ++
Sbjct 41 TSSPTIVLRHMYPNPPVAVAIAEGQNVSD------ELLDQA--------ADHFEAFFSEV 86
Query 71 RSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVC 130
E KYG +ED+V+ D +G V++ Y D +A +A L G RF + +
Sbjct 87 FEELAKYGEVEDMVVCDNIGDHI----IGNVYVKYTDEEAANKALAALQG-RFYSGKQIH 141
Query 131 AAFFPEERFKEG 142
A F P F+E
Sbjct 142 AEFTPVTDFREA 153
> mmu:109095 Rbm15b, 1810017N16Rik; RNA binding motif protein
15B; K13190 RNA-binding protein 15
Length=887
Score = 35.0 bits (79), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 4/59 (6%)
Query 63 HEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGR 121
H + ++R EKYG +E+VVI RP + G FL + ++ A RA++ ++GR
Sbjct 342 HSVSEVELRRAFEKYGIIEEVVIKRP----ARGQGGAYAFLKFQNLDMAHRAKVAMSGR 396
> hsa:29890 RBM15B, HUMAGCGB, OTT3; RNA binding motif protein
15B; K13190 RNA-binding protein 15
Length=890
Score = 35.0 bits (79), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 4/59 (6%)
Query 63 HEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGR 121
H + ++R EKYG +E+VVI RP + G FL + ++ A RA++ ++GR
Sbjct 346 HSVSEVELRRAFEKYGIIEEVVIKRP----ARGQGGAYAFLKFQNLDMAHRAKVAMSGR 400
> cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA binding
protein with 3x RRM domains ; K12838 poly(U)-binding-splicing
factor PUF60
Length=693
Score = 34.7 bits (78), Expect = 0.11, Method: Composition-based stats.
Identities = 29/117 (24%), Positives = 53/117 (45%), Gaps = 6/117 (5%)
Query 23 LSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLED 82
LS+ Q+ + + I+ L N V P+++ + E ++ E KYG + D
Sbjct 577 LSNGPAPYQIPGAISGSASSTNIILLTNMVGPDEIDDELKEE-----VKIECSKYGKVYD 631
Query 83 VVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERF 139
V I N ++S ++F+V+ + A+ A LN R F +V C+ + E +
Sbjct 632 VRIHVSN-NISKPSDRVRIFVVFESPSMAQIAVPALNNRWFGGNQVFCSLYNTERYY 687
> mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U
binding splicing factor 60; K12838 poly(U)-binding-splicing
factor PUF60
Length=499
Score = 33.9 bits (76), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 47/101 (46%), Gaps = 13/101 (12%)
Query 43 SRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPN----EDLSYKPGV 98
S ++ L N V P+D+ D E + E K+G + V+I + ED +
Sbjct 401 STVMVLRNMVDPKDIDDDLEGE-----VTEECGKFGAVNRVIIYQEKQGEEEDAEI---I 452
Query 99 GKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERF 139
K+F+ + + +A LNGR F +VV A + +ERF
Sbjct 453 VKIFVEFSMASETHKAIQALNGRWFGGRKVV-AEVYDQERF 492
> cel:F58B3.7 hypothetical protein; K12840 splicing factor 45
Length=371
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 31/121 (25%), Positives = 51/121 (42%), Gaps = 25/121 (20%)
Query 32 VRAGRTIGEKP----------------SRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAE 75
VR G + E P ++I+QL N +V + + +I+ E E
Sbjct 246 VRGGNIVAEAPKAPTFATNSMEAVQNATKILQLWNLTDLSEVSGEEGKKEFADEIKEEME 305
Query 76 KYGPLEDVVI---PRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAA 132
K G + +V++ ED + VF+ + + A +A +M+NGR F R V A
Sbjct 306 KCGQVVNVIVHVDESQEEDRQVR-----VFVEFTNNAQAIKAFVMMNGRFFG-GRSVSAG 359
Query 133 F 133
F
Sbjct 360 F 360
> hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding
splicing factor 60KDa; K12838 poly(U)-binding-splicing factor
PUF60
Length=516
Score = 33.1 bits (74), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 47/101 (46%), Gaps = 13/101 (12%)
Query 43 SRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPN----EDLSYKPGV 98
S ++ L N V P+D+ D E + E K+G + V+I + ED +
Sbjct 418 STVMVLRNMVDPKDIDDDLEGE-----VTEECGKFGAVNRVIIYQEKQGEEEDAEI---I 469
Query 99 GKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERF 139
K+F+ + + +A LNGR F +VV A + +ERF
Sbjct 470 VKIFVEFSIASETHKAIQALNGRWFAGRKVV-AEVYDQERF 509
> pfa:PF07_0066 conserved Plasmodium protein, unknown function
Length=1125
Score = 33.1 bits (74), Expect = 0.32, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 35/59 (59%), Gaps = 4/59 (6%)
Query 81 EDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERF 139
E+ + PN DL+ +G F+ + ++ SA +A+ L+GR+F ++ A +F E++F
Sbjct 1033 EEEDLTHPNYDLT---SIGCAFIHFENIESATKARKELSGRKFG-ANIIEANYFSEKKF 1087
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIP 86
P++++ L E++ + ++ V+DI+ E +KYG +VV+P
Sbjct 890 PTKVIVLNKIATFEELSDSSEYKDIVEDIKIECDKYGKTLEVVLP 934
> dre:555764 gon4l, fb99g07, udu, wu:fb99g07; gon-4-like (C.elegans)
Length=2055
Score = 33.1 bits (74), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query 42 PSRIVQLLNTVYPEDVMTDASHEAAVKDIRSEAEKYGPLEDVVIPRPNED 91
P+R Q V+PEDV T EA V + S +G E+V +P +ED
Sbjct 1726 PARPGQFEEAVWPEDVATGTDGEAGVS-VTSRGGVWGGFEEVTLPDLDED 1774
> bbo:BBOV_IV007810 23.m06352; U2 splicing factor subunit; K12836
splicing factor U2AF 35 kDa subunit
Length=251
Score = 32.7 bits (73), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 55/122 (45%), Gaps = 20/122 (16%)
Query 22 VLSDPVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDVMTDASH-EAAVKDIRSEAEKYGPL 80
+ +P VAI + G+ I + +LL+ A H E +++ E KYG +
Sbjct 51 MYQNPPVAIAIAEGQMISD------ELLDKA--------ADHFEEFYEEVFLELMKYGEI 96
Query 81 EDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFK 140
ED+V+ D +G V++ Y D SA A ML+GR + + C + P F+
Sbjct 97 EDMVVCDNIGDHI----IGNVYVKYRDENSAAHAISMLSGRFYGGKPIQC-EYTPVTDFR 151
Query 141 EG 142
E
Sbjct 152 EA 153
> tgo:TGME49_112530 splicing factor protein, putative ; K13091
RNA-binding protein 39
Length=633
Score = 32.3 bits (72), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 27/55 (49%), Gaps = 7/55 (12%)
Query 69 DIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRF 123
D+R E +K+G +E V I N D G V++ + AR A LNGR F
Sbjct 566 DVRDECKKFGSVEKVWIDERNVD-------GNVWIRFAHPDQARAAFGALNGRYF 613
> dre:100330844 mCG19533-like; K13190 RNA-binding protein 15
Length=815
Score = 32.3 bits (72), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 63 HEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGR 121
H ++R +KYG +E+VVI RP + G FL + ++ A RA++ + GR
Sbjct 278 HNITEVELRQGFDKYGIIEEVVIKRP----ARGQGGAYAFLKFQNLDMAHRAKVAMQGR 332
> dre:100136872 rbm15b, zgc:175165; RNA binding motif protein
15B
Length=402
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 30/53 (56%), Gaps = 4/53 (7%)
Query 69 DIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGR 121
++R +KYG +E+VVI RP + G FL + ++ A RA++ + GR
Sbjct 56 ELRQGFDKYGIIEEVVIKRP----ARGQGGAYAFLKFQNLDMAHRAKVAMQGR 104
> hsa:7633 ZNF79, pT7; zinc finger protein 79; K09228 KRAB domain-containing
zinc finger protein
Length=498
Score = 32.0 bits (71), Expect = 0.78, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Query 80 LEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRR--FDQTRVVCAAFFPEE 137
LE+ V+P P L + G+ + G +T+ R + F Q R C P +
Sbjct 2 LEEGVLPSPGPALPQEENTGEEGMAAGLLTAGPRGSTFFSSVTVAFAQERWRCLVSTPRD 61
Query 138 RFKEG 142
RFKEG
Sbjct 62 RFKEG 66
> xla:733282 rbm17; RNA binding motif protein 17
Length=400
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 37/71 (52%), Gaps = 11/71 (15%)
Query 74 AEKYGPLEDVVI----PRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRFDQTRVV 129
EKYG + VI P+E+ ++FL + V SA +A + LNGR F R+V
Sbjct 331 CEKYGKVAKCVIFEIPGAPDEEAV------RIFLEFERVESAIKAVVDLNGRYFG-GRIV 383
Query 130 CAAFFPEERFK 140
A F+ ++F+
Sbjct 384 KAGFYNLDKFR 394
> sce:YKL035W UGP1; UDP-glucose pyrophosphorylase (UGPase), catalyses
the reversible formation of UDP-Glc from glucose 1-phosphate
and UTP, involved in a wide variety of metabolic pathways,
expression modulated by Pho85p through Pho4p (EC:2.7.7.9);
K00963 UTP--glucose-1-phosphate uridylyltransferase [EC:2.7.7.9]
Length=499
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 30/99 (30%), Positives = 40/99 (40%), Gaps = 22/99 (22%)
Query 26 PVVAIQVRAGRTIGEKPSRIVQLLNTVYPEDV--------MTDASHEAAVKD-------I 70
P I+VR G T + R ++ LN Y DV TD E +K I
Sbjct 122 PKSVIEVREGNTFLDLSVRQIEYLNRQYDSDVPLLLMNSFNTDKDTEHLIKKYSANRIRI 181
Query 71 RSEAEKYGP--LEDVVIPRPNE-----DLSYKPGVGKVF 102
RS + P +D ++P P E D Y PG G +F
Sbjct 182 RSFNQSRFPRVYKDSLLPVPTEYDSPLDAWYPPGHGDLF 220
> pfa:PFI0820c RNA binding protein, putative
Length=388
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 36/75 (48%), Gaps = 8/75 (10%)
Query 60 DASHEAAVKDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRA-QLML 118
+ S + V +RS EKYG LE+ VI NE S G G FL + +S R A ++M
Sbjct 230 NLSQKTNVSTLRSIFEKYGKLEECVIIHDNEGKS--KGYG--FLTF---SSPREAFKVMQ 282
Query 119 NGRRFDQTRVVCAAF 133
R RVV F
Sbjct 283 QPERIIDNRVVFLHF 297
> xla:447356 MGC130690; MGC84102 protein; K12840 splicing factor
45
Length=400
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query 100 KVFLVYGDVTSARRAQLMLNGRRFDQTRVVCAAFFPEERFK 140
++FL + V SA +A + LNGR F R+V A+F+ ++F+
Sbjct 355 RIFLEFERVESAIKAVVDLNGRYFG-GRIVKASFYNLDKFR 394
> ath:AT3G55460 SCL30; SCL30; RNA binding / nucleic acid binding
/ nucleotide binding
Length=262
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 28/56 (50%), Gaps = 3/56 (5%)
Query 68 KDIRSEAEKYGPLEDVVIPRPNEDLSYKPGVGKVFLVYGDVTSARRAQLMLNGRRF 123
+++R E++GP+ DV IPR D G F+ + D A AQ +N R F
Sbjct 61 EELREPFERFGPVRDVYIPR---DYYSGQPRGFAFVEFVDAYDAGEAQRSMNRRSF 113
Lambda K H
0.317 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40