bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0872_orf1
Length=323
Score E
Sequences producing significant alignments: (Bits) Value
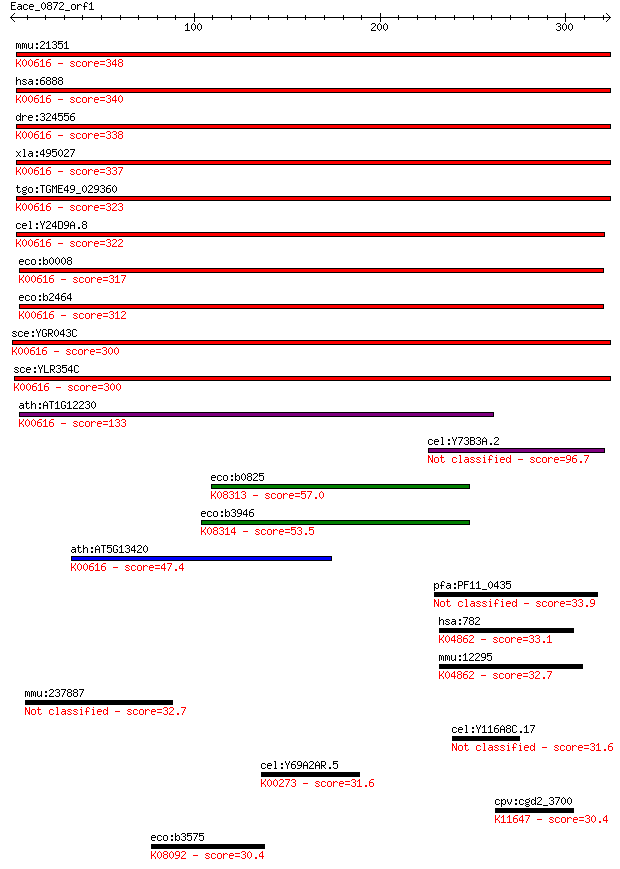
mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transal... 348 2e-95
hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC... 340 3e-93
dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:6... 338 2e-92
xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transa... 337 3e-92
tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616... 323 6e-88
cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.... 322 2e-87
eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2... 317 5e-86
eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);... 312 1e-84
sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC... 300 3e-81
sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC... 300 4e-81
ath:AT1G12230 transaldolase, putative; K00616 transaldolase [E... 133 8e-31
cel:Y73B3A.2 hypothetical protein 96.7 1e-19
eco:b0825 fsaA, ECK0815, JW5109, mipB, ybiZ; fructose-6-phosph... 57.0 9e-08
eco:b3946 fsaB, ECK3938, JW3918, talC, yijG; fructose-6-phosph... 53.5 1e-06
ath:AT5G13420 transaldolase, putative; K00616 transaldolase [E... 47.4 7e-05
pfa:PF11_0435 conserved Plasmodium membrane protein 33.9 0.85
hsa:782 CACNB1, CAB1, CACNLB1, CCHLB1, MGC41896; calcium chann... 33.1 1.4
mmu:12295 Cacnb1, Cchb1, Cchlb1; calcium channel, voltage-depe... 32.7 1.7
mmu:237887 Slfn10-ps, FLJ00257, Slfn10, mFLJ00257; schlafen 10... 32.7 1.8
cel:Y116A8C.17 dct-13; DAF-16/FOXO Controlled, germline Tumor ... 31.6 3.9
cel:Y69A2AR.5 hypothetical protein; K00273 D-amino-acid oxidas... 31.6 4.5
cpv:cgd2_3700 SWI/SNF related transcriptional regulator ATpase... 30.4 9.1
eco:b3575 yiaK, ECK3564, JW3547; 2,3-diketo-L-gulonate reducta... 30.4 9.2
> mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=337
Score = 348 bits (892), Expect = 2e-95, Method: Compositional matrix adjust.
Identities = 172/322 (53%), Positives = 236/322 (73%), Gaps = 6/322 (1%)
Query 4 SALAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEA 63
SAL QL + +T+VAD+ DF AI E+ P DATTNP+L+L A P Y+ ++E+A+ K+
Sbjct 13 SALDQLKQFTTVVADTGDFNAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGKKL 72
Query 64 TKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYE 123
G P + +++A+D++ V FG E+L+ IPG VSTE+ A LSFD++ V RARR+I LY+
Sbjct 73 --GGPQEEQIKNAIDKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYK 130
Query 124 EKGISRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFV 182
E G+ ++RILIK+++TWEGIQA K+L+E++ I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 131 EAGVGKDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFV 190
Query 183 GRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGC 242
GRILDW+ K S+ +DPGVKSV +IYNY+K FG KT VM ASFRNT EI LAGC
Sbjct 191 GRILDWHVANTDKKSYEPQEDPGVKSVTKIYNYYKKFGYKTIVMGASFRNTGEIKALAGC 250
Query 243 DKLTISPKLLEELNKLKDK-EVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKL 301
D LTISPKLL EL LKD +++ LSV+ A +K+ ++EK FRW+ NED MA EKL
Sbjct 251 DFLTISPKLLGEL--LKDNSKLAPALSVKAAQTSDSEKIHLDEKAFRWLHNEDQMAVEKL 308
Query 302 AEGIRNFSADMNKLKELIKQHM 323
++GIR F+AD KL+ ++ + M
Sbjct 309 SDGIRKFAADAIKLERMLTERM 330
> hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 340 bits (873), Expect = 3e-93, Method: Compositional matrix adjust.
Identities = 169/321 (52%), Positives = 232/321 (72%), Gaps = 4/321 (1%)
Query 4 SALAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEA 63
SAL QL + +T+VAD+ DF AI E+ P DATTNP+L+L A P Y+ ++E+A+ ++
Sbjct 13 SALDQLKQFTTVVADTGDFHAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGRKL 72
Query 64 TKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYE 123
G + +++A+D++ V FG E+L+ IPG VSTE+ A LSFD++ V RARR+I LY+
Sbjct 73 --GGSQEDQIKNAIDKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYK 130
Query 124 EKGISRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFV 182
E GIS++RILIK+++TWEGIQA K+L+E++ I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 131 EAGISKDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFV 190
Query 183 GRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGC 242
GRILDW+ K S+ +DPGVKSV +IYNY+K F KT VM ASFRNT EI LAGC
Sbjct 191 GRILDWHVANTDKKSYEPLEDPGVKSVTKIYNYYKKFSYKTIVMGASFRNTGEIKALAGC 250
Query 243 DKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKLA 302
D LTISPKLL EL + K V LS + A ++K+ ++EK FRW+ NED MA EKL+
Sbjct 251 DFLTISPKLLGELLQDNAKLVPV-LSAKAAQASDLEKIHLDEKSFRWLHNEDQMAVEKLS 309
Query 303 EGIRNFSADMNKLKELIKQHM 323
+GIR F+AD KL+ ++ + M
Sbjct 310 DGIRKFAADAVKLERMLTERM 330
> dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:66054;
transaldolase 1 (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 338 bits (867), Expect = 2e-92, Method: Compositional matrix adjust.
Identities = 165/321 (51%), Positives = 231/321 (71%), Gaps = 4/321 (1%)
Query 4 SALAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEA 63
SAL Q+ K + +VAD+ DF AI+E+ P DATTNP+L+L A P Y+ ++++A+K
Sbjct 13 SALEQIKKFTVVVADTGDFNAIEEYKPQDATTNPSLILAAAKMPAYQPLVDQAIKYG--T 70
Query 64 TKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYE 123
G D V +A+D++ V FG+E+L+ +PG VSTE+ A LSFD++ V RARR+ISLYE
Sbjct 71 ANGGTEDEQVTNAMDKLFVNFGLEILKKVPGRVSTEVDARLSFDKDAMVSRARRLISLYE 130
Query 124 EKGISRERILIKIAATWEGIQAAKQLKE-ENINCNITLLFSLCQAIAASDAGAYLVSPFV 182
+ GIS+ERILIK+++TWEGIQA ++L+E I+CN+TLLFS QA+A ++A L+SPFV
Sbjct 131 DAGISKERILIKLSSTWEGIQAGRELEEIHGIHCNMTLLFSFAQAVACAEARVTLISPFV 190
Query 183 GRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGC 242
GRILDWYK + ++ +DPGV SV +IYNY+K F +T VM ASFRN E+ LAGC
Sbjct 191 GRILDWYKENTERKTYEPHEDPGVLSVTKIYNYYKKFDYRTVVMGASFRNIGEVKALAGC 250
Query 243 DKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKLA 302
D LTISP LLEEL++ ++ L+ Q A + +++L ++EK FRW+ NED MA EKL+
Sbjct 251 DLLTISPALLEELSQ-DHSAITCTLTPQTARECVLERLQLDEKSFRWLHNEDRMAVEKLS 309
Query 303 EGIRNFSADMNKLKELIKQHM 323
+GIR F+AD KL+ +IK+ M
Sbjct 310 DGIRKFAADAVKLETMIKERM 330
> xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=338
Score = 337 bits (864), Expect = 3e-92, Method: Compositional matrix adjust.
Identities = 165/321 (51%), Positives = 233/321 (72%), Gaps = 4/321 (1%)
Query 4 SALAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEA 63
SAL QL + + +VAD+ DF AI+E+ P DATTNP+L+L A P Y+ ++ A++ K
Sbjct 14 SALDQLKQHTVVVADTGDFNAIEEYKPQDATTNPSLILAAAQMPDYQGLVNDAIQYGKNL 73
Query 64 TKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYE 123
G + + + +D++ V FG+E+L+ IPG VSTE+ A LSFD++G VERA+R+I+LY+
Sbjct 74 --GGSEEEQINNIMDKLFVLFGVEILKKIPGRVSTEVDARLSFDKDGMVERAKRLIALYK 131
Query 124 EKGISRERILIKIAATWEGIQAAKQLKEE-NINCNITLLFSLCQAIAASDAGAYLVSPFV 182
E GI ++RILIK+++TWEGIQA K L+EE I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 132 EAGIDKKRILIKLSSTWEGIQAGKILEEEYGIHCNMTLLFSFAQAVACAEAGVTLISPFV 191
Query 183 GRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGC 242
GRILDW+ S+ ++DPGVKSV +IYNY+K FG KT VM ASFRNT EI L GC
Sbjct 192 GRILDWHVANSDNKSYEPSEDPGVKSVAKIYNYYKKFGYKTIVMGASFRNTGEIKALTGC 251
Query 243 DKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKLA 302
D LTISPKLL EL K + +++ L+V+ A ++K+ + EK+FRW+ NED MA EKL+
Sbjct 252 DYLTISPKLLSELAK-DNSKLTPALTVKEAQASNLEKVHLEEKEFRWLHNEDQMAVEKLS 310
Query 303 EGIRNFSADMNKLKELIKQHM 323
+GIR F+ D KL++++K+ +
Sbjct 311 DGIRKFAIDAIKLEKMLKERL 331
> tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616
transaldolase [EC:2.2.1.2]
Length=364
Score = 323 bits (828), Expect = 6e-88, Method: Compositional matrix adjust.
Identities = 176/330 (53%), Positives = 249/330 (75%), Gaps = 13/330 (3%)
Query 4 SALAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEA 63
+AL L LS +VAD+ DF A+K++ P DATTNP+L+L+A P+Y H++++AV+ AK+
Sbjct 35 NALEALRLLSVVVADTGDFVALKKYVPHDATTNPSLLLKAASMPQYAHLMDEAVEVAKKV 94
Query 64 -TKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLY 122
K +P D VE+ +D++ V FG+E+L+I+PG+VSTE+ A LSFD + SVE+AR++I +Y
Sbjct 95 HGKVEPDDELVEEVIDQLFVRFGVEILKIVPGVVSTEVSAALSFDVQASVEKARKLILMY 154
Query 123 EEKGISRERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFV 182
+E GI ++R+LIK+A TWEG +AAK L++E I+CN+TLLFS QA+AA++A A L+SPFV
Sbjct 155 KEHGIEKDRVLIKLATTWEGCEAAKILEKEGIHCNMTLLFSFAQAVAAAEAKATLISPFV 214
Query 183 GRILDWYKLKDPK--ASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLA 240
GRILD+YK P+ A + GA DPGV+SV++IY Y+K KT VMAASFRN EI+ LA
Sbjct 215 GRILDYYKKLHPEKVAEYVGAQDPGVQSVKRIYKYYKKHNYKTVVMAASFRNIGEIIALA 274
Query 241 GCDKLTISPKLLEELNKLKDKEVSTRL-------SVQLAAKEKIQKLDINEKKFRWMLNE 293
GCD++T+SP LLEE LK+ ++ R SV+ + E +KL+++EK FRWMLNE
Sbjct 275 GCDRVTVSPALLEE---LKNSDLPVRRVLGEPTESVEASDAEDEKKLEMDEKTFRWMLNE 331
Query 294 DAMATEKLAEGIRNFSADMNKLKELIKQHM 323
DAMATEKLAEGIR+F+ D+ LKE+I++ +
Sbjct 332 DAMATEKLAEGIRSFNRDLLSLKEMIREKL 361
> cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.2.1.2]
Length=319
Score = 322 bits (824), Expect = 2e-87, Method: Compositional matrix adjust.
Identities = 165/318 (51%), Positives = 227/318 (71%), Gaps = 4/318 (1%)
Query 4 SALAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEA 63
S L QL S +VAD+ DF AIKEF PTDATTNP+L+L A +Y +++++V AKE
Sbjct 2 SVLEQLKGASVVVADTGDFNAIKEFQPTDATTNPSLILAASKMEQYAALIDQSVAYAKEH 61
Query 64 TKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYE 123
G ++ A+DR+ V FG E+L+ IPG VSTE+ A LSFD + S++RA +I+ YE
Sbjct 62 ASGHQE--VLQAAMDRLFVVFGKEILKTIPGRVSTEVDARLSFDTQASIDRALGLIAQYE 119
Query 124 EKGISRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFV 182
++GIS++RILIK+A+TWEGI+AAK L+ ++ I+CN+TLLF+ QA+A +++G L+SPFV
Sbjct 120 KEGISKDRILIKLASTWEGIRAAKFLESKHGIHCNMTLLFNFEQAVACAESGVTLISPFV 179
Query 183 GRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGC 242
GRI+DW+ + +++ DDPGV SV +I+NY+K + KT VMAASFRNT+EI L GC
Sbjct 180 GRIMDWFVKNTDQKAYTRKDDPGVVSVTRIFNYYKKYDYKTQVMAASFRNTEEIKGLVGC 239
Query 243 DKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKLA 302
D LTISP LL++L + + LS A + K+ I+EK FRW LNEDAMATEKLA
Sbjct 240 DLLTISPALLKQLAA-ETELAPVVLSTSHAKTLDLPKVSIDEKAFRWALNEDAMATEKLA 298
Query 303 EGIRNFSADMNKLKELIK 320
EGIRNF+ D L++LI+
Sbjct 299 EGIRNFAKDARTLEKLIE 316
> eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=317
Score = 317 bits (811), Expect = 5e-86, Method: Compositional matrix adjust.
Identities = 152/314 (48%), Positives = 218/314 (69%), Gaps = 5/314 (1%)
Query 6 LAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEATK 65
L L + +T+VAD+ D A+K + P DATTNP+L+L A P+Y+ +++ AV AK+ +
Sbjct 5 LTSLRQYTTVVADTGDIAAMKLYQPQDATTNPSLILNAAQIPEYRKLIDDAVAWAKQQSN 64
Query 66 GKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYEEK 125
+ + DA D++ V G+E+L+++PG +STE+ A LS+D E S+ +A+R+I LY +
Sbjct 65 DRAQQIV--DATDKLAVNIGLEILKLVPGRISTEVDARLSYDTEASIAKAKRLIKLYNDA 122
Query 126 GISRERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRI 185
GIS +RILIK+A+TW+GI+AA+QL++E INCN+TLLFS QA A ++AG +L+SPFVGRI
Sbjct 123 GISNDRILIKLASTWQGIRAAEQLEKEGINCNLTLLFSFAQARACAEAGVFLISPFVGRI 182
Query 186 LDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGCDKL 245
LDWYK K ++ A+DPGV SV +IY Y+K G +T VM ASFRN EIL LAGCD+L
Sbjct 183 LDWYKANTDKKEYAPAEDPGVVSVSEIYQYYKEHGYETVVMGASFRNIGEILELAGCDRL 242
Query 246 TISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKLAEGI 305
TI+P LL+E L + E + + + K + I E +F W N+D MA +KLAEGI
Sbjct 243 TIAPALLKE---LAESEGAIERKLSYTGEVKARPARITESEFLWQHNQDPMAVDKLAEGI 299
Query 306 RNFSADMNKLKELI 319
R F+ D KL+++I
Sbjct 300 RKFAIDQEKLEKMI 313
> eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=316
Score = 312 bits (799), Expect = 1e-84, Method: Compositional matrix adjust.
Identities = 151/315 (47%), Positives = 224/315 (71%), Gaps = 7/315 (2%)
Query 6 LAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAKEATK 65
L + + +T+VADS D E+I+ + P DATTNP+L+L+A +Y+H+++ A+ K+
Sbjct 4 LDGIKQFTTVVADSGDIESIRHYHPQDATTNPSLLLKAAGLSQYEHLIDDAIAWGKK--N 61
Query 66 GKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISLYEEK 125
GK + V A D++ V FG E+L+I+PG VSTE+ A LSFD+E S+E+AR ++ LY+++
Sbjct 62 GKTQEQQVVAACDKLAVNFGAEILKIVPGRVSTEVDARLSFDKEKSIEKARHLVDLYQQQ 121
Query 126 GISRERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRI 185
G+ + RILIK+A+TWEGI+AA++L++E INCN+TLLFS QA A ++AG +L+SPFVGRI
Sbjct 122 GVEKSRILIKLASTWEGIRAAEELEKEGINCNLTLLFSFAQARACAEAGVFLISPFVGRI 181
Query 186 LDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLAGCDKL 245
DWY+ + P + +DPGVKSVR IY+Y+K +T VM ASFR T++IL L GCD+L
Sbjct 182 YDWYQARKPMDPYVVEEDPGVKSVRNIYDYYKQHHYETIVMGASFRRTEQILALTGCDRL 241
Query 246 TISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQK-LDINEKKFRWMLNEDAMATEKLAEG 304
TI+P LL+EL ++VS + + + + ++E +FRW N+DAMA EKL+EG
Sbjct 242 TIAPNLLKELQ----EKVSPVVRKLIPPSQTFPRPAPMSEAEFRWEHNQDAMAVEKLSEG 297
Query 305 IRNFSADMNKLKELI 319
IR F+ D KL++L+
Sbjct 298 IRLFAVDQRKLEDLL 312
> sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=333
Score = 300 bits (769), Expect = 3e-81, Method: Compositional matrix adjust.
Identities = 155/326 (47%), Positives = 231/326 (70%), Gaps = 8/326 (2%)
Query 2 GSSALAQLAKLST-IVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEA 60
+S+L QL K T +VADS DFEAI ++ P D+TTNP+L+L A KY ++ AV+
Sbjct 12 ATSSLEQLKKAGTHVVADSGDFEAISKYEPQDSTTNPSLILAASKLEKYARFIDAAVEYG 71
Query 61 KEATKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIIS 120
++ GK +E+A+D++LV FG ++L+++PG VSTE+ A LSFD++ +V++A II
Sbjct 72 RK--HGKTDHEKIENAMDKILVEFGTQILKVVPGRVSTEVDARLSFDKKATVKKALHIIK 129
Query 121 LYEEKGISRERILIKIAATWEGIQAAKQLK-EENINCNITLLFSLCQAIAASDAGAYLVS 179
LY++ G+ +ER+LIKIA+TWEGIQAA++L+ + I+CN+TLLFS QA+A ++A L+S
Sbjct 130 LYKDAGVPKERVLIKIASTWEGIQAARELEVKHGIHCNMTLLFSFTQAVACAEANVTLIS 189
Query 180 PFVGRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHL 239
PFVGRI+D+YK K ++ DPGV SV++IY+Y+K G T VMAASFRN DE+ L
Sbjct 190 PFVGRIMDFYKALSGK-DYTAETDPGVLSVKKIYSYYKRHGYATEVMAASFRNLDELKAL 248
Query 240 AGCDKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDI--NEKKFRWMLNEDAMA 297
AG D +T+ LLE+L + D + +L+ + A +E ++K+ +E FR++LNED MA
Sbjct 249 AGIDNMTLPLNLLEQLYESTDP-IENKLNSESAKEEGVEKVSFINDEPHFRYVLNEDQMA 307
Query 298 TEKLAEGIRNFSADMNKLKELIKQHM 323
TEKL++GIR FSAD+ L +L+++ M
Sbjct 308 TEKLSDGIRKFSADIEALYKLVEEKM 333
> sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=335
Score = 300 bits (769), Expect = 4e-81, Method: Compositional matrix adjust.
Identities = 172/325 (52%), Positives = 238/325 (73%), Gaps = 8/325 (2%)
Query 3 SSALAQL-AKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPKYKHVLEKAVKEAK 61
+++L QL A + +VAD+ DF +I +F P D+TTNP+L+L A P Y +++ AV+ K
Sbjct 13 NNSLEQLKASGTVVVADTGDFGSIAKFQPQDSTTNPSLILAAAKQPTYAKLIDVAVEYGK 72
Query 62 EATKGKPADVTVEDAVDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGSVERARRIISL 121
+ GK + VE+AVDR+LV FG E+L+I+PG VSTE+ A LSFD + ++E+AR II L
Sbjct 73 K--HGKTTEEQVENAVDRLLVEFGKEILKIVPGRVSTEVDARLSFDTQATIEKARHIIKL 130
Query 122 YEEKGISRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSP 180
+E++G+S+ER+LIKIA+TWEGIQAAK+L+E++ I+CN+TLLFS QA+A ++A L+SP
Sbjct 131 FEQEGVSKERVLIKIASTWEGIQAAKELEEKDGIHCNLTLLFSFVQAVACAEAQVTLISP 190
Query 181 FVGRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRNTDEILHLA 240
FVGRILDWYK K + G DPGV SV++IYNY+K +G KT VM ASFR+TDEI +LA
Sbjct 191 FVGRILDWYKSSTGK-DYKGEADPGVISVKKIYNYYKKYGYKTIVMGASFRSTDEIKNLA 249
Query 241 GCDKLTISPKLLEELNKLKD--KEVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMAT 298
G D LTISP LL++L + V +S + A +KI + +E KFR+ LNEDAMAT
Sbjct 250 GVDYLTISPALLDKLMNSTEPFPRVLDPVSAKKEAGDKISYIS-DESKFRFDLNEDAMAT 308
Query 299 EKLAEGIRNFSADMNKLKELIKQHM 323
EKL+EGIR FSAD+ L +LI++ +
Sbjct 309 EKLSEGIRKFSADIVTLFDLIEKKV 333
> ath:AT1G12230 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=405
Score = 133 bits (335), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 89/268 (33%), Positives = 150/268 (55%), Gaps = 24/268 (8%)
Query 6 LAQLAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPK--YKHVLEKAVKEAKEA 63
L ++ S IV D+ F+ + F PT AT + L+L P +++ ++ A+ ++ A
Sbjct 71 LNAVSAFSEIVPDTVVFDDFERFPPTAATVSSALLLGICGLPDTIFRNAVDMALADSSCA 130
Query 64 TKGKPADVTVEDAVDRVLVFF-------GMELLEIIPGLVSTEIPADLSFDQEGSVERAR 116
+E R+ FF G +L++++PG VSTE+ A L++D G + +
Sbjct 131 G--------LETTESRLSCFFNKAIVNVGGDLVKLVPGRVSTEVDARLAYDTNGIIRKVH 182
Query 117 RIISLYEEKGISRERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAY 176
++ LY E + +R+L KI ATW+GI+AA+ L+ E I ++T ++S QA AAS AGA
Sbjct 183 DLLRLYNEIDVPHDRLLFKIPATWQGIEAARLLESEGIQTHMTFVYSFAQAAAASQAGAS 242
Query 177 LVSPFVGRILDWYKLK----DPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAASFRN 232
++ FVGR+ DW + + +++ +DPG+ V++ YNY +G K+ +MAA+ RN
Sbjct 243 VIQIFVGRLRDWARNHSGDTEIESAIKSGEDPGLALVKRSYNYIHKYGYKSKLMAAAVRN 302
Query 233 TDEILHLAGCDKLTISPKLLEELNKLKD 260
++ L G D + I+P L+ L LKD
Sbjct 303 KQDLFSLLGVDYV-IAP--LKVLQSLKD 327
> cel:Y73B3A.2 hypothetical protein
Length=97
Score = 96.7 bits (239), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 53/95 (55%), Positives = 66/95 (69%), Gaps = 1/95 (1%)
Query 226 MAASFRNTDEILHLAGCDKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEK 285
MAASFRNT+EI L GCD LTISP LL++L + + LS A + K+ I+EK
Sbjct 1 MAASFRNTEEIKGLVGCDLLTISPALLKQLAA-ETEFAPVVLSTSHAKTLDLPKVSIDEK 59
Query 286 KFRWMLNEDAMATEKLAEGIRNFSADMNKLKELIK 320
FRW LN+DAMATEKLAEGIRNF+ D L++LI+
Sbjct 60 AFRWALNDDAMATEKLAEGIRNFAKDARTLEKLIE 94
> eco:b0825 fsaA, ECK0815, JW5109, mipB, ybiZ; fructose-6-phosphate
aldolase 1 (EC:4.1.2.-); K08313 fructose-6-phosphate aldolase
1 [EC:4.1.2.-]
Length=220
Score = 57.0 bits (136), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 67/141 (47%), Gaps = 22/141 (15%)
Query 109 EGSVERARRIISLYEEKGISRERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAI 168
EG V A ++ S+ + I++K+ T EG+ A K LK E I T ++ Q +
Sbjct 66 EGMVNDALKLRSIIAD-------IVVKVPVTAEGLAAIKMLKAEGIPTLGTAVYGAAQGL 118
Query 169 AASDAGAYLVSPFVGRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKTTVMAA 228
++ AGA V+P+V RI G+++V ++ K + V+AA
Sbjct 119 LSALAGAEYVAPYVNRI-------------DAQGGSGIQTVTDLHQLLKMHAPQAKVLAA 165
Query 229 SFRNTDEILH--LAGCDKLTI 247
SF+ + L LAGC+ +T+
Sbjct 166 SFKTPRQALDCLLAGCESITL 186
> eco:b3946 fsaB, ECK3938, JW3918, talC, yijG; fructose-6-phosphate
aldolase 2 (EC:4.1.2.-); K08314 fructose-6-phosphate aldolase
2 [EC:4.1.2.-]
Length=220
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/146 (30%), Positives = 76/146 (52%), Gaps = 22/146 (15%)
Query 104 LSFDQEGSVERARRIISLYEEKGISRERILIKIAATWEGIQAAKQLKEENINCNITLLFS 163
+S D +G VE A+R+ + I++KI T EG+ A K LK+E I T ++S
Sbjct 61 MSRDAQGMVEEAKRLRD-------AIPGIVVKIPVTSEGLAAIKILKKEGITTLGTAVYS 113
Query 164 LCQAIAASDAGAYLVSPFVGRILDWYKLKDPKASFSGADDPGVKSVRQIYNYFKHFGSKT 223
Q + A+ AGA V+P+V R+ G D G+++V+++ + ++
Sbjct 114 AAQGLLAALAGAKYVAPYVNRV-----------DAQGGD--GIRTVQELQTLLEMHAPES 160
Query 224 TVMAASFRNTDEILH--LAGCDKLTI 247
V+AASF+ + L LAGC+ +T+
Sbjct 161 MVLAASFKTPRQALDCLLAGCESITL 186
> ath:AT5G13420 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=438
Score = 47.4 bits (111), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 38/141 (26%), Positives = 70/141 (49%), Gaps = 6/141 (4%)
Query 34 TTNPTLVLQAVHNPKYKHVLEKAVKEAKEATKGKPADVTVEDAVDRVLVFFGM-ELLEII 92
T+NP + +A+ + + + E+ + + ++ V+D D +F + + E
Sbjct 112 TSNPAIFQKAISTSNAYNDQFRTLVESGKDIESAYWELVVKDIQDACKLFEPIYDQTEGA 171
Query 93 PGLVSTEIPADLSFDQEGSVERARRIISLYEEKGISRERILIKIAATWEGIQAAKQLKEE 152
G VS E+ L+ D +G+VE A+ Y K ++R + IKI AT I + + +
Sbjct 172 DGYVSVEVSPRLADDTQGTVEAAK-----YLSKVVNRRNVYIKIPATAPCIPSIRDVIAA 226
Query 153 NINCNITLLFSLCQAIAASDA 173
I+ N+TL+FS+ + A DA
Sbjct 227 GISVNVTLIFSIARYEAVIDA 247
> pfa:PF11_0435 conserved Plasmodium membrane protein
Length=1828
Score = 33.9 bits (76), Expect = 0.85, Method: Composition-based stats.
Identities = 24/89 (26%), Positives = 42/89 (47%), Gaps = 1/89 (1%)
Query 229 SFRNTDEILHLA-GCDKLTISPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLDINEKKF 287
S TD +L+L+ G + ++ L E L ++ + V KE+ K D++EKK
Sbjct 933 SVEKTDILLNLSNGKNNGNVTSSLCENLFVYNQDKIQRKKKVPYKNKERDNKDDLDEKKD 992
Query 288 RWMLNEDAMATEKLAEGIRNFSADMNKLK 316
++ N+D+ +G+ MNK K
Sbjct 993 MYICNDDSSVITSSEKGVTKERIHMNKEK 1021
> hsa:782 CACNB1, CAB1, CACNLB1, CCHLB1, MGC41896; calcium channel,
voltage-dependent, beta 1 subunit; K04862 voltage-dependent
calcium channel beta-1
Length=598
Score = 33.1 bits (74), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 43/84 (51%), Gaps = 14/84 (16%)
Query 232 NTDEILHLAGCDKLTI----------SPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLD 281
+ D I H A K ++ SPK+L+ L K + K S L+VQ+AA EK+ +
Sbjct 322 DADTINHPAQLSKTSLAPIIVYIKITSPKVLQRLIKSRGKSQSKHLNVQIAASEKLAQCP 381
Query 282 INEKKFRWMLNEDAM--ATEKLAE 303
+ F +L+E+ + A E LAE
Sbjct 382 --PEMFDIILDENQLEDACEHLAE 403
> mmu:12295 Cacnb1, Cchb1, Cchlb1; calcium channel, voltage-dependent,
beta 1 subunit; K04862 voltage-dependent calcium channel
beta-1
Length=479
Score = 32.7 bits (73), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 45/89 (50%), Gaps = 14/89 (15%)
Query 232 NTDEILHLAGCDKLTI----------SPKLLEELNKLKDKEVSTRLSVQLAAKEKIQKLD 281
+ D I H A K ++ SPK+L+ L K + K S L+VQ+AA EK+ +
Sbjct 322 DADTINHPAQLSKTSLAPIIVYIKITSPKVLQRLIKSRGKSQSKHLNVQIAASEKLAQCP 381
Query 282 INEKKFRWMLNEDAM--ATEKLAEGIRNF 308
+ F +L+E+ + A E LAE + +
Sbjct 382 --PEMFDIILDENQLEDACEHLAEYLEAY 408
> mmu:237887 Slfn10-ps, FLJ00257, Slfn10, mFLJ00257; schlafen
10, pseudogene
Length=847
Score = 32.7 bits (73), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 38/81 (46%), Gaps = 7/81 (8%)
Query 9 LAKLSTIVADSADFEAIKEFSPTDATTNPTLVLQAVHNPK--YKHVLEKAVKEAKEATKG 66
L K+ + VAD D K +SP D VL + K Y+H+ + +++ K A++
Sbjct 723 LEKMVSYVADKCDVFLSKGYSPQDIA-----VLFSTDREKKAYEHMFLREIRKRKRASQM 777
Query 67 KPADVTVEDAVDRVLVFFGME 87
A V + D + F G+E
Sbjct 778 NDASVCHSNMFDSIRRFSGLE 798
> cel:Y116A8C.17 dct-13; DAF-16/FOXO Controlled, germline Tumor
affecting family member (dct-13)
Length=205
Score = 31.6 bits (70), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 239 LAGCDKLTISPKLLEELNKLKDKEVSTRLSVQLAAK 274
+A CD TIS +L EE+ +LK KE + + S+ L+ K
Sbjct 67 IANCDPCTISDELREEMMRLKKKEKAFKTSLCLSHK 102
> cel:Y69A2AR.5 hypothetical protein; K00273 D-amino-acid oxidase
[EC:1.4.3.3]
Length=322
Score = 31.6 bits (70), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 5/58 (8%)
Query 136 IAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPF-----VGRILDW 188
+ A GI +A ++E NC +T++ SD A L+ PF V RI++W
Sbjct 7 LGAGINGIASALAIQERLPNCEVTIIAEKFSPNTTSDVAAGLIEPFLCDDDVDRIINW 64
> cpv:cgd2_3700 SWI/SNF related transcriptional regulator ATpase
; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1552
Score = 30.4 bits (67), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 262 EVSTRLSVQLAAKEKIQKLDINEKKFRWMLNEDAMATEKLAE 303
EV+ L ++ +I+ LD +E+K RW N+D + E L+E
Sbjct 1225 EVNAELGIEDNNLNEIRDLDRSERKSRWKSNKDYLCVEGLSE 1266
> eco:b3575 yiaK, ECK3564, JW3547; 2,3-diketo-L-gulonate reductase,
NADH-dependent (EC:1.1.1.-); K08092 3-dehydro-L-gulonate
2-dehydrogenase [EC:1.1.1.130]
Length=332
Score = 30.4 bits (67), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 32/68 (47%), Gaps = 8/68 (11%)
Query 77 VDRVLVFFGMELLEIIPGLVSTEIPADLSFDQEGS-------VERARRIISLYEEKGISR 129
VD + F +LE+ L ++P D FD EG+ +E+ RRI+ + KG
Sbjct 171 VDMSMSMFSYGMLEV-NRLAGRQLPVDGGFDDEGNLTKEPGVIEKNRRILPMGYWKGSGM 229
Query 130 ERILIKIA 137
+L IA
Sbjct 230 SIVLDMIA 237
Lambda K H
0.316 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13331501400
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40