bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0887_orf4
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
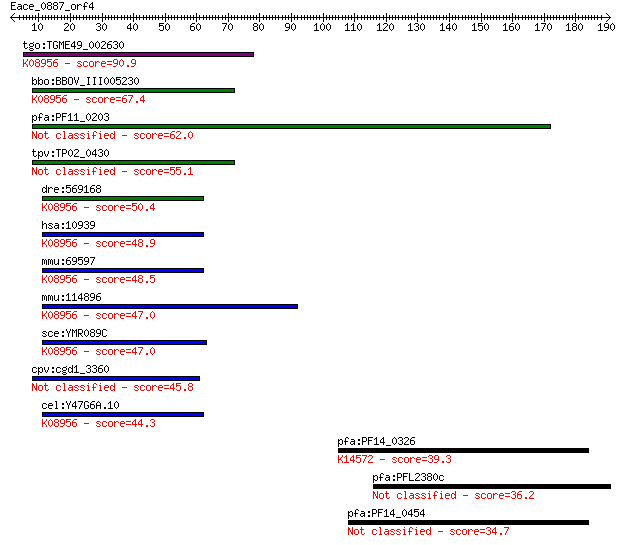
tgo:TGME49_002630 AFG3 ATPase family protein ; K08956 AFG3 fam... 90.9 3e-18
bbo:BBOV_III005230 17.m07468; ATP-dependent metalloprotease Ft... 67.4 3e-11
pfa:PF11_0203 peptidase, putative 62.0 1e-09
tpv:TP02_0430 hypothetical protein 55.1 1e-07
dre:569168 si:ch211-12e1.4; K08956 AFG3 family protein [EC:3.4... 50.4 4e-06
hsa:10939 AFG3L2, FLJ25993, SCA28; AFG3 ATPase family gene 3-l... 48.9 1e-05
mmu:69597 Afg3l2, 2310036I02Rik, AW260507, Emv66, par; AFG3(AT... 48.5 1e-05
mmu:114896 Afg3l1, 1700047G05Rik, 3110061K15Rik; AFG3(ATPase f... 47.0 4e-05
sce:YMR089C YTA12, RCA1; Component, with Afg3p, of the mitocho... 47.0 4e-05
cpv:cgd1_3360 AFG1 ATpase family AAA ATpase 45.8 8e-05
cel:Y47G6A.10 spg-7; human SPG (spastic paraplegia) family mem... 44.3 3e-04
pfa:PF14_0326 dynein-related AAA-type ATPase; K14572 midasin 39.3 0.009
pfa:PFL2380c leucine-rich repeat protein 12, LRR12 36.2 0.065
pfa:PF14_0454 conserved Plasmodium protein, unknown function 34.7 0.20
> tgo:TGME49_002630 AFG3 ATPase family protein ; K08956 AFG3 family
protein [EC:3.4.24.-]
Length=1188
Score = 90.9 bits (224), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 62/84 (73%), Gaps = 11/84 (13%)
Query 5 QDPYQLFSDATAQLVDDEVRNLISNQYERVKALLKEKEK-----------KETLTFADLQ 53
Q+ Y+ +S+ TA+++DDEV +I++QYERVK LLKE+EK +E++T++++
Sbjct 1018 QNFYRPYSEHTAKVIDDEVSQIINDQYERVKTLLKEREKEVHSLCELLISRESITYSEIL 1077
Query 54 DCLGTRPFPPDAQLAAYINALPTK 77
+C+G RP PPD Q+AAYI ALPT+
Sbjct 1078 ECIGPRPVPPDPQMAAYIQALPTR 1101
> bbo:BBOV_III005230 17.m07468; ATP-dependent metalloprotease
FtsH family protein; K08956 AFG3 family protein [EC:3.4.24.-]
Length=797
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/75 (40%), Positives = 48/75 (64%), Gaps = 11/75 (14%)
Query 8 YQLFSDATAQLVDDEVRNLISNQYERVKALLKEK-----------EKKETLTFADLQDCL 56
Y+ +S+ TAQL+D EVR +I +QY RVK++L+EK ++ET+T+ D+ C+
Sbjct 680 YRTYSENTAQLIDTEVRTMIESQYARVKSMLREKAELVHKLSKLLYQRETITYHDIASCI 739
Query 57 GTRPFPPDAQLAAYI 71
G R FP + +L Y+
Sbjct 740 GEREFPVEEKLRPYV 754
> pfa:PF11_0203 peptidase, putative
Length=1052
Score = 62.0 bits (149), Expect = 1e-09, Method: Composition-based stats.
Identities = 52/177 (29%), Positives = 85/177 (48%), Gaps = 22/177 (12%)
Query 8 YQLFSDATAQLVDDEVRNLISNQYERVKALLKEKEK-----------KETLTFADLQDCL 56
Y+ S+ A L+D+EVR+LI QY+RVK++L + EK KET+++ D+ C+
Sbjct 852 YRPHSECLAHLIDNEVRSLIETQYKRVKSILMKNEKHVHNLANLLYEKETISYHDIVKCV 911
Query 57 GTRPFPPDAQLAAYINALPTKVNTVGQEEPGVE-RASGDLRRIPTAISGDRNVRD-EKGS 114
G RP+P + ++ A P K + EP +E + D + T+ G+ N D EKG
Sbjct 912 GERPYPVKSAYEKFVKANPYKAIS---SEPLLEDKKESDNMKGDTSNIGNVNTNDFEKGH 968
Query 115 TSDKDAAADGHSMQQRKQSIKDTEGSNGDDKEPTDAGKNGGKGKKKRNGKGNAADDD 171
+ + + K++ KD N + N K + N GN++ DD
Sbjct 969 IKENTKEC---TKENTKENTKDNTKENTKENTKEYTKDN---TKNQYNILGNSSKDD 1019
> tpv:TP02_0430 hypothetical protein
Length=881
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 43/75 (57%), Gaps = 11/75 (14%)
Query 8 YQLFSDATAQLVDDEVRNLISNQYERVKALLKEKEK-----------KETLTFADLQDCL 56
Y+ +S+ TAQL+D +VR +I +QY RVK +L K + KET+T+ D+ C+
Sbjct 801 YRNYSETTAQLIDQQVRTIIEDQYLRVKNMLLGKAELVHKLSKLLYDKETITYHDIVQCV 860
Query 57 GTRPFPPDAQLAAYI 71
G R FP + YI
Sbjct 861 GEREFPMKDRYKPYI 875
> dre:569168 si:ch211-12e1.4; K08956 AFG3 family protein [EC:3.4.24.-]
Length=800
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 38/62 (61%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
+S+ATA+L+D EVRNLIS YER + LL +K+ +KE L D+ + LG R
Sbjct 684 YSEATARLIDTEVRNLISTAYERTQQLLSDKKPEVEKVALRLLEKEVLDKNDMVELLGKR 743
Query 60 PF 61
PF
Sbjct 744 PF 745
> hsa:10939 AFG3L2, FLJ25993, SCA28; AFG3 ATPase family gene 3-like
2 (S. cerevisiae); K08956 AFG3 family protein [EC:3.4.24.-]
Length=797
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
+S+ATA+L+DDEVR LI++ Y+R ALL EK+ +KE L D+ + LG R
Sbjct 689 YSEATARLIDDEVRILINDAYKRTVALLTEKKADVEKVALLLLEKEVLDKNDMVELLGPR 748
Query 60 PF 61
PF
Sbjct 749 PF 750
> mmu:69597 Afg3l2, 2310036I02Rik, AW260507, Emv66, par; AFG3(ATPase
family gene 3)-like 2 (yeast); K08956 AFG3 family protein
[EC:3.4.24.-]
Length=802
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 37/62 (59%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
+S+ATA+++DDEVR LIS+ Y R ALL EK+ +KE L D+ LG R
Sbjct 688 YSEATARMIDDEVRILISDAYRRTVALLTEKKADVEKVALLLLEKEVLDKNDMVQLLGPR 747
Query 60 PF 61
PF
Sbjct 748 PF 749
> mmu:114896 Afg3l1, 1700047G05Rik, 3110061K15Rik; AFG3(ATPase
family gene 3)-like 1 (yeast); K08956 AFG3 family protein [EC:3.4.24.-]
Length=789
Score = 47.0 bits (110), Expect = 4e-05, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 51/103 (49%), Gaps = 22/103 (21%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALL----KEKEK-------KETLTFADLQDCLGTR 59
+S+ATAQL+D+EVR L+ + Y R LL ++ EK KE L AD+ + LG R
Sbjct 681 YSEATAQLIDEEVRCLVRSAYNRTLELLTQCREQVEKVGRRLLEKEVLEKADMIELLGPR 740
Query 60 PFPPDAQLAAYI---------NALPTKVN--TVGQEEPGVERA 91
PF + ++ +LP + G+EE G ER
Sbjct 741 PFAEKSTYEEFVEGTGSLEEDTSLPEGLKDWNKGREEGGTERG 783
> sce:YMR089C YTA12, RCA1; Component, with Afg3p, of the mitochondrial
inner membrane m-AAA protease that mediates degradation
of misfolded or unassembled proteins and is also required
for correct assembly of mitochondrial enzyme complexes (EC:3.4.24.-);
K08956 AFG3 family protein [EC:3.4.24.-]
Length=825
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 33/63 (52%), Gaps = 11/63 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
FSD T ++D EV ++ ++R LLKEK KKE LT D+ D LG R
Sbjct 727 FSDETGDIIDSEVYRIVQECHDRCTKLLKEKAEDVEKIAQVLLKKEVLTREDMIDLLGKR 786
Query 60 PFP 62
PFP
Sbjct 787 PFP 789
> cpv:cgd1_3360 AFG1 ATpase family AAA ATpase
Length=719
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 40/64 (62%), Gaps = 11/64 (17%)
Query 8 YQLFSDATAQLVDDEVRNLISNQYERVKALLKEKEK-----------KETLTFADLQDCL 56
Y+ +S+AT+Q +D+ +R +I++QY RVK LL K++ KET+T D+ +C+
Sbjct 653 YKPYSEATSQAIDNCIRKMINDQYSRVKELLILKKEQVHKLSDLLLNKETVTNQDINECI 712
Query 57 GTRP 60
G P
Sbjct 713 GPMP 716
> cel:Y47G6A.10 spg-7; human SPG (spastic paraplegia) family member
(spg-7); K08956 AFG3 family protein [EC:3.4.24.-]
Length=782
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 35/62 (56%), Gaps = 11/62 (17%)
Query 11 FSDATAQLVDDEVRNLISNQYERVKALLKEKE-----------KKETLTFADLQDCLGTR 59
+S+ATAQL+D EVR+L+ N R + LL EK +KE L D+ + +G R
Sbjct 674 YSEATAQLIDQEVRDLVMNALRRTRDLLLEKRSDIERVALRLLEKEILNREDMIELVGKR 733
Query 60 PF 61
PF
Sbjct 734 PF 735
> pfa:PF14_0326 dynein-related AAA-type ATPase; K14572 midasin
Length=8105
Score = 39.3 bits (90), Expect = 0.009, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 4/81 (4%)
Query 105 DRNVRDEKGSTSDKDAAADGHSMQQRKQSIKDTEGSNGDDKEPTDAGKNGGKGKKKRNGK 164
D++ D G DKD A G Q+ + IK+ D E D + K +K +
Sbjct 7214 DKSYSDNDGEKEDKDDA--GQDNQKWEDDIKNENKDKHDKNEKNDKNEKNDKNEKNDKNE 7271
Query 165 GNAADDDDDDNDDDD--DDRN 183
N ++ +D NDDD DD N
Sbjct 7272 KNDKNEKNDKNDDDVQLDDNN 7292
> pfa:PFL2380c leucine-rich repeat protein 12, LRR12
Length=863
Score = 36.2 bits (82), Expect = 0.065, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Query 116 SDKDAAADGHSMQQRKQSIKDTEGSNGDDK-EPTDAGKNGGKGKKKRNGKGNAADDDDDD 174
SDK D K+ D + +GD+K E D K G KK+ G+ +D D+
Sbjct 546 SDKKGDIDNEKFGHNKKEDGDNKKEDGDNKKEDGDNKKEDGDNKKE---DGDNKKEDGDN 602
Query 175 NDDDDDDRNGNNSKRG 190
+ D+++NG+N K G
Sbjct 603 EKNGDNEKNGDNKKNG 618
> pfa:PF14_0454 conserved Plasmodium protein, unknown function
Length=1986
Score = 34.7 bits (78), Expect = 0.20, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 34/76 (44%), Gaps = 8/76 (10%)
Query 108 VRDEKGSTSDKDAAADGHSMQQRKQSIKDTEGSNGDDKEPTDAGKNGGKGKKKRNGKGNA 167
+ D+KG D+ DG D + N D ++ D GKN K K GK +
Sbjct 171 IDDDKGENDDRGKNDDGGK--------NDDKEKNDDKEKNYDKGKNDDKEKNYNKGKNDD 222
Query 168 ADDDDDDNDDDDDDRN 183
+ +DD +DD ++N
Sbjct 223 KEKNDDKEKNDDKEKN 238
Lambda K H
0.305 0.128 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5429324208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40