bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0897_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
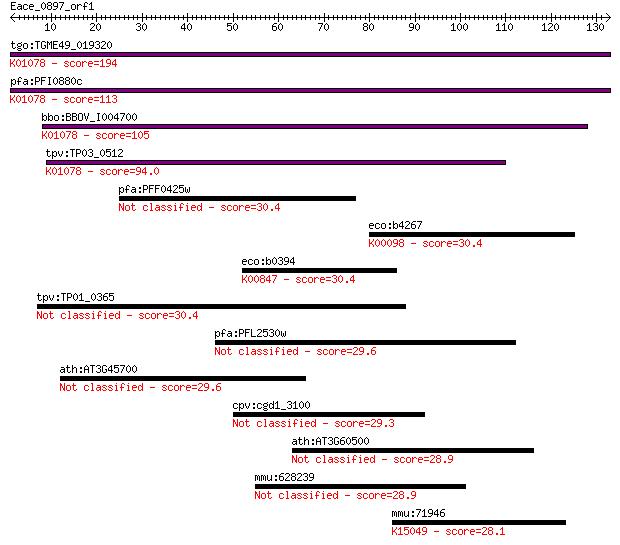
tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01... 194 6e-50
pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3... 113 2e-25
bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01... 105 4e-23
tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosp... 94.0 1e-19
pfa:PFF0425w conserved Plasmodium protein, unknown function 30.4 1.3
eco:b4267 idnD, ECK4260, JW4224, yjgV; L-idonate 5-dehydrogena... 30.4 1.3
eco:b0394 mak, ECK0389, JW0385, yajF; manno(fructo)kinase (EC:... 30.4 1.7
tpv:TP01_0365 hypothetical protein 30.4 1.7
pfa:PFL2530w lysophospholipase, putative 29.6 2.4
ath:AT3G45700 proton-dependent oligopeptide transport (POT) fa... 29.6 2.5
cpv:cgd1_3100 hypothetical protein 29.3 3.6
ath:AT3G60500 CER7; CER7 (ECERIFERUM 7); 3'-5'-exoribonuclease... 28.9 4.1
mmu:628239 zinc finger protein 14-like 28.9 4.3
mmu:71946 Endod1, 2210414F18Rik, 2310067E08Rik, C85344; endonu... 28.1 6.9
> tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=431
Score = 194 bits (493), Expect = 6e-50, Method: Compositional matrix adjust.
Identities = 84/132 (63%), Positives = 109/132 (82%), Gaps = 0/132 (0%)
Query 1 DDDISYINCGTGSLSGGSALVKASGSKFFSGDKGFCLFEMSAEGLVTKFINGENGETLYE 60
DD+IS+++CG GS + GS +VK SGS +++G+ GFCLFE++AEGLVT+ ++G GETLY
Sbjct 300 DDNISHVSCGAGSKAAGSPIVKHSGSLYYAGETGFCLFELTAEGLVTRLVSGTTGETLYT 359
Query 61 HKQPLKNRPERSTIDQFNYFTDMPEVTYFPVPEMGKLPGKDVFVRVVGTIGLCILTFLGT 120
HKQPLKNRPER +ID FN+ + +PEV Y+PVPEMGK+PG+DVFVRVVGTIGLCI T +
Sbjct 360 HKQPLKNRPERKSIDAFNFVSQLPEVRYYPVPEMGKMPGRDVFVRVVGTIGLCIATIFLS 419
Query 121 LGIATGISRAMK 132
L +A G+SR MK
Sbjct 420 LSVANGLSRYMK 431
> pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3.2);
K01078 acid phosphatase [EC:3.1.3.2]
Length=396
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/132 (40%), Positives = 84/132 (63%), Gaps = 0/132 (0%)
Query 1 DDDISYINCGTGSLSGGSALVKASGSKFFSGDKGFCLFEMSAEGLVTKFINGENGETLYE 60
D+D+++I CG+GS+S G + +K S S FFS D GFC+ E+S G+VTKF++ + GE +Y
Sbjct 265 DNDMAHITCGSGSMSQGKSGMKNSKSLFFSSDIGFCVHELSNNGIVTKFVSSKKGEVIYT 324
Query 61 HKQPLKNRPERSTIDQFNYFTDMPEVTYFPVPEMGKLPGKDVFVRVVGTIGLCILTFLGT 120
HK +K + ++ +F +P V VP G + KD FVRVVGTIG+ I + +
Sbjct 325 HKLNIKKKKTLDKVNALQHFAALPNVELTDVPSSGPMGNKDTFVRVVGTIGILIGSVIVF 384
Query 121 LGIATGISRAMK 132
+G ++ +S+ MK
Sbjct 385 IGASSFLSKNMK 396
> bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=395
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 55/126 (43%), Positives = 76/126 (60%), Gaps = 9/126 (7%)
Query 8 NCGTGSLSGGSALVKASGSKFFSGDKGFCLFEMSAEGLVTKFINGENGETLYEHKQPLKN 67
N GTG G ++K + S FFS GFC+ E+ A+G+ TKFINGE G+ +Y HKQ +K
Sbjct 273 NAGTG---GRKPIMKTTNSAFFSEKAGFCIHELGADGMETKFINGETGDVMYTHKQAIKK 329
Query 68 RPERSTIDQFNYFTDMPEVTYFPVPEMGKLPGKDVFVRVVGTIGLCI----LTFLG--TL 121
RP+R ++ + + +P V+ +P+ EM D FV++VGTIGL I LT L TL
Sbjct 330 RPQRQYGNEVQHVSALPTVSLYPIGEMASPTQMDAFVKIVGTIGLIIAGLHLTLLSGTTL 389
Query 122 GIATGI 127
G A I
Sbjct 390 GKAASI 395
> tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosphatase
[EC:3.1.3.2]
Length=404
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 41/101 (40%), Positives = 61/101 (60%), Gaps = 0/101 (0%)
Query 9 CGTGSLSGGSALVKASGSKFFSGDKGFCLFEMSAEGLVTKFINGENGETLYEHKQPLKNR 68
CG+ G +++K+ SKF++ GFCL E++AEG TKF+NG GE LY H QP KNR
Sbjct 283 CGSSGNKGRKSVIKSPHSKFYTEAPGFCLHELNAEGFTTKFVNGNTGEVLYTHVQPKKNR 342
Query 69 PERSTIDQFNYFTDMPEVTYFPVPEMGKLPGKDVFVRVVGT 109
+R + +PEVT+ P+ ++ + D F ++VGT
Sbjct 343 KQRQHGSELKLINKLPEVTFHPLGDLEGVSYSDAFTKIVGT 383
> pfa:PFF0425w conserved Plasmodium protein, unknown function
Length=283
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query 25 GSKFFSGDKGFCLFEMSAEGLVT---KFINGENGETLYEHKQPLKNRPERSTIDQ 76
G+K ++GF ++M+ E LV K+ N + G+ K KNR E ID+
Sbjct 125 GTKNLEENEGFSQYKMNQEDLVNDNKKYSNEKYGDMTNHSKNSFKNRNEHMMIDE 179
> eco:b4267 idnD, ECK4260, JW4224, yjgV; L-idonate 5-dehydrogenase,
NAD-binding (EC:1.1.1.264); K00098 L-idonate 5-dehydrogenase
[EC:1.1.1.264]
Length=343
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 80 FTDMPEVTYFPVPEMGKLPGKDVFVRVVGTIGLCILTFLGTLGIA 124
F + V + G+L GK VF+ VG IG I++ + TLG A
Sbjct 151 FAEPLAVAIHAAHQAGELQGKRVFISGVGPIGCLIVSAVKTLGAA 195
> eco:b0394 mak, ECK0389, JW0385, yajF; manno(fructo)kinase (EC:2.7.1.4);
K00847 fructokinase [EC:2.7.1.4]
Length=302
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 16/34 (47%), Gaps = 0/34 (0%)
Query 52 GENGETLYEHKQPLKNRPERSTIDQFNYFTDMPE 85
G+ GE LY H+ P R TI+ DM E
Sbjct 18 GDAGEQLYRHRLPTPRDDYRQTIETIATLVDMAE 51
> tpv:TP01_0365 hypothetical protein
Length=296
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 40/82 (48%), Gaps = 1/82 (1%)
Query 7 INCGTGSLSGGSALVKASGSKFFSGDK-GFCLFEMSAEGLVTKFINGENGETLYEHKQPL 65
IN T +S G ++ + S+ + + + EM++ G T +N E+GET + +P+
Sbjct 10 INTQTHHISQGRSVRVSEPSERVTNRRMATQVIEMNSRGYPTDRVNFESGETTHTSVEPV 69
Query 66 KNRPERSTIDQFNYFTDMPEVT 87
R+T+ +Y + VT
Sbjct 70 DLNSNRTTLSPRSYDPENNSVT 91
> pfa:PFL2530w lysophospholipase, putative
Length=453
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Query 46 VTKFINGENGETLYEHKQPLKNRPERSTID-QFNYFTDMPEVTYFPVPEMGKLPGKDVFV 104
V ++IN N + E + P E S D +N+ MP + P+ MG G ++ +
Sbjct 132 VIQYINKINSSVIKEREDP----KEYSYSDYNYNFKNKMPNIVRSPLYIMGLSMGGNIAL 187
Query 105 RVVGTIG 111
RV+ IG
Sbjct 188 RVLELIG 194
> ath:AT3G45700 proton-dependent oligopeptide transport (POT)
family protein
Length=548
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 30/54 (55%), Gaps = 5/54 (9%)
Query 12 GSLSGGSALVKASGSKFFSGDKGFCLFEMSAEGLVTKFINGENGETLYEHKQPL 65
G+++G +A+V A + + G C+ A L++ FI G+ LYEH QPL
Sbjct 183 GAITGTTAIVYAQDNASWKLGFGLCV----AANLIS-FIIFVAGKRLYEHDQPL 231
> cpv:cgd1_3100 hypothetical protein
Length=610
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 50 INGENGETLYEHKQPLKNRPERSTIDQFNYFTDMPEVTYFPV 91
+N N L +P KN+P+ S ID +++ ++FP+
Sbjct 1 MNKNNNVCLGRRGRPKKNQPKPSLIDCIEALSEIDNFSFFPI 42
> ath:AT3G60500 CER7; CER7 (ECERIFERUM 7); 3'-5'-exoribonuclease/
RNA binding
Length=438
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 30/60 (50%), Gaps = 7/60 (11%)
Query 63 QPLKNRPERSTIDQFNYFTDMPEVTYFP------VPEMGKLPGKDVF-VRVVGTIGLCIL 115
QP K+RP ++ F F+ M + ++ P E+G++ + + R V T LC+L
Sbjct 70 QPYKDRPNEGSLSIFTEFSPMADPSFEPGRPGESAVELGRIIDRGLRESRAVDTESLCVL 129
> mmu:628239 zinc finger protein 14-like
Length=462
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query 55 GETLYEHKQPLKNRPERSTIDQFNYFTDMPEVTYFPVPEMGKLPGK 100
GE LYE Q K P+RST+ ++ +T + E Y+ V + G+ P K
Sbjct 365 GEKLYEFNQCSKAFPKRSTLG-YHKWTHIGE-KYYEVNQFGEKPFK 408
> mmu:71946 Endod1, 2210414F18Rik, 2310067E08Rik, C85344; endonuclease
domain containing 1; K15049 endonuclease domain-containing
1 protein
Length=501
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 85 EVTYFPVPEMGKLP--GKDVFVRVVGTIGLCILTFLGTLG 122
+V FPV +G +P KD+ V + GT+ L T GT+G
Sbjct 437 DVATFPVYTVGAIPIVCKDIAVGLGGTLSLLFDTAFGTVG 476
Lambda K H
0.319 0.140 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40