bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0902_orf1
Length=102
Score E
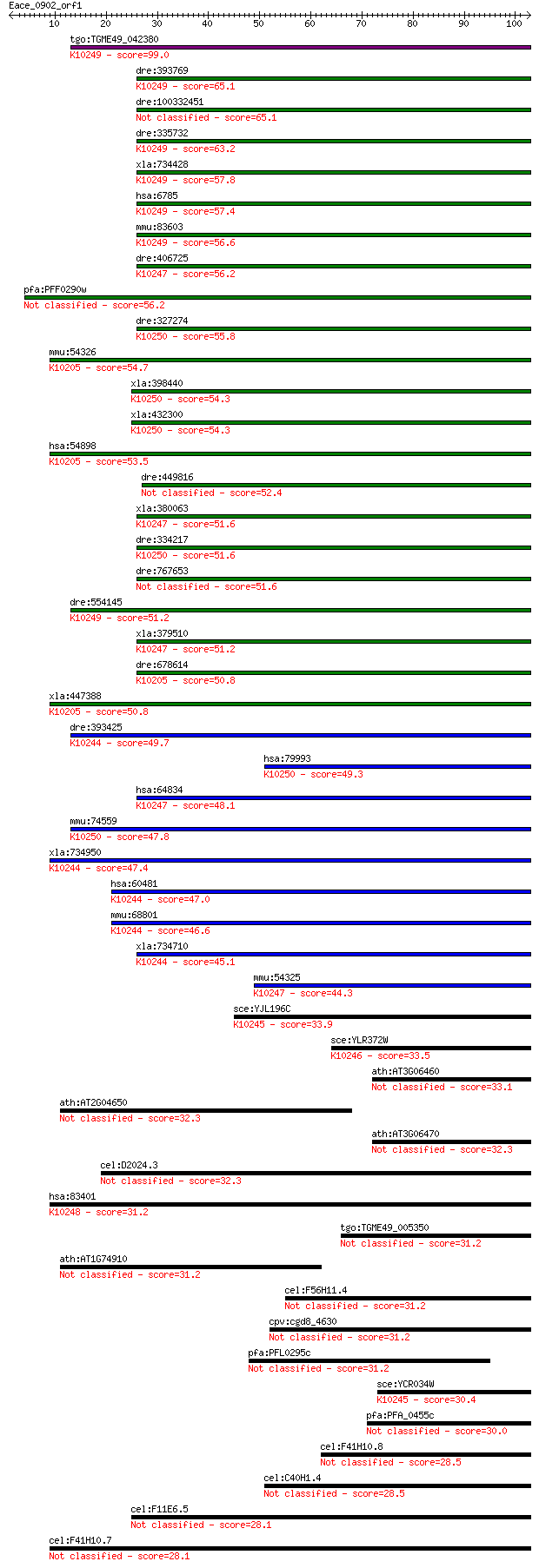
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_042380 elongation of very long chain fatty acids 4 ... 99.0 3e-21
dre:393769 elovl4a, MGC73341, zgc:73341; elongation of very lo... 65.1 5e-11
dre:100332451 elongation of very long chain fatty acids-like 4... 65.1 6e-11
dre:335732 elovl4b, elovl4, wu:fk62b04, zgc:73054; elongation ... 63.2 2e-10
xla:734428 hypothetical protein MGC115163; K10249 elongation o... 57.8 8e-09
hsa:6785 ELOVL4, ADMD, CT118, FLJ17667, FLJ92876, STGD2, STGD3... 57.4 1e-08
mmu:83603 Elovl4; elongation of very long chain fatty acids (F... 56.6 2e-08
dre:406725 elovl1b, wu:fj32h05, zgc:56567; elongation of very ... 56.2 2e-08
pfa:PFF0290w long chain polyunsaturated fatty acid elongation ... 56.2 2e-08
dre:327274 elovl7b, elovl1, wu:fd20a06, zgc:56422; ELOVL famil... 55.8 3e-08
mmu:54326 Elovl2, AI317360, Ssc2; elongation of very long chai... 54.7 7e-08
xla:398440 elovl7; ELOVL family member 7, elongation of long c... 54.3 9e-08
xla:432300 hypothetical protein MGC80262; K10250 elongation of... 54.3 1e-07
hsa:54898 ELOVL2, FLJ20334, SSC2; elongation of very long chai... 53.5 2e-07
dre:449816 elovl1a, zgc:103538; elongation of very long chain ... 52.4 4e-07
xla:380063 elovl1, MGC52731; elongation of very long chain fat... 51.6 5e-07
dre:334217 elovl7a, MGC174838, elovl7, fi36f02, wu:fi36f02, zg... 51.6 6e-07
dre:767653 MGC153394; zgc:153394 51.6 7e-07
dre:554145 im:7139490; zgc:112263; K10249 elongation of very l... 51.2 7e-07
xla:379510 hypothetical protein MGC64517; K10247 elongation of... 51.2 8e-07
dre:678614 elovl2, MGC136352, MGC158526, zgc:136352; elongatio... 50.8 1e-06
xla:447388 elovl2, MGC84669; elongation of very long chain fat... 50.8 1e-06
dre:393425 elovl5, MGC63549, zgc:63549; ELOVL family member 5,... 49.7 2e-06
hsa:79993 ELOVL7, FLJ23563; ELOVL family member 7, elongation ... 49.3 3e-06
hsa:64834 ELOVL1, Ssc1; elongation of very long chain fatty ac... 48.1 6e-06
mmu:74559 Elovl7, 9130013K24Rik, AI840082; ELOVL family member... 47.8 8e-06
xla:734950 elovl5, MGC131143; ELOVL family member 5, elongatio... 47.4 1e-05
hsa:60481 ELOVL5, HELO1, dJ483K16.1; ELOVL family member 5, el... 47.0 1e-05
mmu:68801 Elovl5, 1110059L23Rik, AI747313, AU043003, HELO1; EL... 46.6 2e-05
xla:734710 hypothetical protein MGC114802; K10244 elongation o... 45.1 5e-05
mmu:54325 Elovl1, AA407424, BB151133, Ssc1; elongation of very... 44.3 8e-05
sce:YJL196C ELO1; Elo1p; K10245 fatty acid elongase 2 [EC:2.3.... 33.9 0.12
sce:YLR372W SUR4, APA1, ELO3, SRE1, VBM1; Elongase, involved i... 33.5 0.16
ath:AT3G06460 GNS1/SUR4 membrane family protein 33.1 0.20
ath:AT2G04650 ADP-glucose pyrophosphorylase family protein 32.3 0.35
ath:AT3G06470 GNS1/SUR4 membrane family protein 32.3 0.36
cel:D2024.3 elo-3; fatty acid ELOngation family member (elo-3) 32.3 0.41
hsa:83401 ELOVL3, CIG-30, CIG30, MGC21435; elongation of very ... 31.2 0.84
tgo:TGME49_005350 fatty acid elongation protein, putative 31.2 0.84
ath:AT1G74910 ADP-glucose pyrophosphorylase family protein 31.2 0.85
cel:F56H11.4 elo-1; fatty acid ELOngation family member (elo-1) 31.2 0.87
cpv:cgd8_4630 7 pass integral membrane proteinwith FLHWFHH mot... 31.2 0.87
pfa:PFL0295c conserved Plasmodium membrane protein 31.2 0.92
sce:YCR034W FEN1, ELO2, GNS1, VBM2; Fatty acid elongase, invol... 30.4 1.5
pfa:PFA_0455c fatty acid elongation protein, GNS1/SUR4 family,... 30.0 1.9
cel:F41H10.8 elo-6; fatty acid ELOngation family member (elo-6) 28.5 5.3
cel:C40H1.4 elo-4; fatty acid ELOngation family member (elo-4) 28.5 6.1
cel:F11E6.5 elo-2; fatty acid ELOngation family member (elo-2) 28.1 6.7
cel:F41H10.7 elo-5; fatty acid ELOngation family member (elo-5) 28.1 7.2
> tgo:TGME49_042380 elongation of very long chain fatty acids
4 protein, putative ; K10249 elongation of very long chain fatty
acids protein 4
Length=350
Score = 99.0 bits (245), Expect = 3e-21, Method: Composition-based stats.
Identities = 43/90 (47%), Positives = 60/90 (66%), Gaps = 0/90 (0%)
Query 13 LLEKMGPVFVCQAIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNTRMAEVTWFF 72
L EKM + + Q +YNI QV+ C ++V + V+ AE Y+ + N+F+ MAE+ W F
Sbjct 122 LAEKMSFILLLQIVYNIFQVVTCSYVVYKAMSVYVAEGYTIVFNKFDPSRRNMAEIVWLF 181
Query 73 YMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
Y+ K VDLLDTVF++ RG W Q SFLH+YH
Sbjct 182 YLTKVVDLLDTVFIVCRGKWAQFSFLHIYH 211
> dre:393769 elovl4a, MGC73341, zgc:73341; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4a;
K10249 elongation of very long chain fatty acids protein 4
Length=309
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 36/81 (44%), Positives = 50/81 (61%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNV---PN-TRMAEVTWFFYMLKYVDLL 81
IYN + VIL F+ +F RA YS+IC + PN R+A W++++ K V+ L
Sbjct 70 IYNFSMVILNFFIFKELFLAARAANYSYICQPVDYSDDPNEVRVAAALWWYFISKGVEYL 129
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF I+R + Q+SFLHVYH
Sbjct 130 DTVFFILRKKFNQISFLHVYH 150
> dre:100332451 elongation of very long chain fatty acids-like
4-like
Length=298
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 50/81 (61%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNV---PN-TRMAEVTWFFYMLKYVDLL 81
IYN + VIL F+ +F RA YS+IC + PN R+A W++++ K V+ L
Sbjct 59 IYNFSMVILNFFIFKELFLAARAANYSYICQPVDYSDDPNEVRVAAALWWYFISKGVEYL 118
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF I+R + Q+SFLHVYH
Sbjct 119 DTVFFILRKKFNQISFLHVYH 139
> dre:335732 elovl4b, elovl4, wu:fk62b04, zgc:73054; elongation
of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
4b; K10249 elongation of very long chain fatty acids
protein 4
Length=303
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 48/81 (59%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPN----TRMAEVTWFFYMLKYVDLL 81
+YN + V+L ++ + RA YS++C N N R+A W++Y+ K V+ L
Sbjct 70 VYNFSMVLLNFYICKELLLGSRAAGYSYLCQPVNYSNDVNEVRIASALWWYYISKGVEFL 129
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF I+R + QVSFLHVYH
Sbjct 130 DTVFFIMRKKFNQVSFLHVYH 150
> xla:734428 hypothetical protein MGC115163; K10249 elongation
of very long chain fatty acids protein 4
Length=265
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 47/81 (58%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
+YN++ V L ++ YS++C + N+ RMA V W+F+ K ++LL
Sbjct 67 VYNLSLVGLSVYMFYEFLVTSVLAGYSYLCQPVDYSNSELGMRMARVCWWFFFSKVIELL 126
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DT+F I+R + Q+SFLHVYH
Sbjct 127 DTIFFIMRKKFNQISFLHVYH 147
> hsa:6785 ELOVL4, ADMD, CT118, FLJ17667, FLJ92876, STGD2, STGD3;
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3,
yeast)-like 4; K10249 elongation of very long chain
fatty acids protein 4
Length=314
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 34/81 (41%), Positives = 45/81 (55%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRF----NVPNTRMAEVTWFFYMLKYVDLL 81
IYN V+L F+ +F YS+IC NV R+A W++++ K V+ L
Sbjct 81 IYNFGMVLLNLFIFRELFMGSYNAGYSYICQSVDYSNNVHEVRIAAALWWYFVSKGVEYL 140
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF I+R QVSFLHVYH
Sbjct 141 DTVFFILRKKNNQVSFLHVYH 161
> mmu:83603 Elovl4; elongation of very long chain fatty acids
(FEN1/Elo2, SUR4/Elo3, yeast)-like 4; K10249 elongation of very
long chain fatty acids protein 4
Length=312
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 45/81 (55%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPN----TRMAEVTWFFYMLKYVDLL 81
IYN V+L F+ +F YS+IC + N R+A W++++ K V+ L
Sbjct 81 IYNFGMVLLNLFIFRELFMGSYNAGYSYICQSVDYSNDVNEVRIAGALWWYFVSKGVEYL 140
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF I+R QVSFLHVYH
Sbjct 141 DTVFFILRKKNNQVSFLHVYH 161
> dre:406725 elovl1b, wu:fj32h05, zgc:56567; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
1b; K10247 elongation of very long chain fatty acids protein
1
Length=320
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 47/81 (58%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
IYN++ V L ++V A Y++ C+ + N+ RMA V W F K+++L+
Sbjct 69 IYNLSLVGLSAYIVYEFLMSGWATGYTWRCDPCDYSNSPQGLRMARVAWLFLFSKFIELM 128
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF ++R Q++FLH++H
Sbjct 129 DTVFFVLRKKHSQITFLHIFH 149
> pfa:PFF0290w long chain polyunsaturated fatty acid elongation
enzyme, putative
Length=293
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/101 (32%), Positives = 53/101 (52%), Gaps = 6/101 (5%)
Query 4 AVTQKREKPLLEKMGPVFVCQAIYNIAQVILCGFLVARVFDVWRAEE--YSFICNRFNVP 61
A+T+ + LE+M V YN+ QV+ L+ + V+ A+ +S N +
Sbjct 80 AITRSHKSTPLEQM--VEKLTPSYNLLQVLFS--LIITLLTVYEAKNRRFSLFYNSVDFS 135
Query 62 NTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+A W FY+ K VD +DT+ +++R W Q +FLHVYH
Sbjct 136 KKNIALCCWLFYLNKLVDFVDTILIVLRKKWNQFTFLHVYH 176
> dre:327274 elovl7b, elovl1, wu:fd20a06, zgc:56422; ELOVL family
member 7, elongation of long chain fatty acids (yeast) b;
K10250 elongation of very long chain fatty acids protein 7
Length=282
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 47/81 (58%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
IYN++ V+ +++ A Y++ C+ + ++ RMA W +Y K++++L
Sbjct 70 IYNLSIVLFSLYMIYEFLMSGWANGYTYRCDLVDYSSSPQALRMAWTCWLYYFSKFIEML 129
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF ++R QVSFLHVYH
Sbjct 130 DTVFFVLRKKSSQVSFLHVYH 150
> mmu:54326 Elovl2, AI317360, Ssc2; elongation of very long chain
fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2; K10205
elongation of very long chain fatty acids protein 2 [EC:2.3.1.-]
Length=292
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 9/98 (9%)
Query 9 REKPLLEKMGPVFVCQAIYNIAQVILCGF-LVARVFDVWRAEEYSFICNRFNVP---NTR 64
+ +P L G + +YN+A +L + LV + W Y+ C + + R
Sbjct 57 KNRPALSLRGIL----TLYNLAITLLSAYMLVELILSSWEGG-YNLQCQNLDSAGEGDVR 111
Query 65 MAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+A+V W++Y K V+ LDT+F ++R Q++FLHVYH
Sbjct 112 VAKVLWWYYFSKLVEFLDTIFFVLRKKTNQITFLHVYH 149
> xla:398440 elovl7; ELOVL family member 7, elongation of long
chain fatty acids; K10250 elongation of very long chain fatty
acids protein 7
Length=302
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 44/82 (53%), Gaps = 4/82 (4%)
Query 25 AIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDL 80
A YN+ V+ ++ A YSF C+ + + RMA W FY K+++L
Sbjct 69 ACYNLFMVLFSVYMCYEFLMSGWATGYSFRCDIVDYSQSPQALRMAWTCWLFYFSKFIEL 128
Query 81 LDTVFMIVRGNWRQVSFLHVYH 102
LDTVF ++R Q++FLHVYH
Sbjct 129 LDTVFFVLRKKNSQITFLHVYH 150
> xla:432300 hypothetical protein MGC80262; K10250 elongation
of very long chain fatty acids protein 7
Length=301
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 44/82 (53%), Gaps = 4/82 (4%)
Query 25 AIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDL 80
A YN+ V+ ++ A YSF C+ + + RMA W FY K+++L
Sbjct 69 ACYNLFMVLFSVYMCYEFLMSGWATGYSFRCDIVDYSRSPQALRMAWTCWLFYFSKFIEL 128
Query 81 LDTVFMIVRGNWRQVSFLHVYH 102
LDTVF ++R Q++FLHVYH
Sbjct 129 LDTVFFVLRKKNSQITFLHVYH 150
> hsa:54898 ELOVL2, FLJ20334, SSC2; elongation of very long chain
fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2; K10205
elongation of very long chain fatty acids protein 2 [EC:2.3.1.-]
Length=296
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 51/98 (52%), Gaps = 9/98 (9%)
Query 9 REKPLLEKMGPVFVCQAIYNIAQVILCGFLVAR-VFDVWRAEEYSFICNRFNVP---NTR 64
+ +P L G + +YN+ +L +++A + W Y+ C + R
Sbjct 57 KNRPALSLRGIL----TLYNLGITLLSAYMLAELILSTWEGG-YNLQCQDLTSAGEADIR 111
Query 65 MAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+A+V W++Y K V+ LDT+F ++R Q++FLHVYH
Sbjct 112 VAKVLWWYYFSKSVEFLDTIFFVLRKKTSQITFLHVYH 149
> dre:449816 elovl1a, zgc:103538; elongation of very long chain
fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1a
Length=315
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 42/77 (54%), Gaps = 8/77 (10%)
Query 27 YNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT-RMAEVTWFFYMLKYVDLLDTVF 85
Y + + ++ G+ WR + +C+ + P RM W FY KY++LLDTVF
Sbjct 80 YTVYEFLMSGWATTYT---WRCD----LCDPSSSPQALRMVRAAWLFYFSKYIELLDTVF 132
Query 86 MIVRGNWRQVSFLHVYH 102
++R QV+FLH++H
Sbjct 133 FVLRKKHSQVTFLHIFH 149
> xla:380063 elovl1, MGC52731; elongation of very long chain fatty
acids (FEN1/Elo2, SUR4/Elo3)-like 1; K10247 elongation
of very long chain fatty acids protein 1
Length=290
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 44/81 (54%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
+YN + V L F+V Y++ C+ +V ++ RM V W F K+++LL
Sbjct 68 VYNFSLVALSAFIVYEFLMSGWLTGYTWRCDPVDVSDSPMALRMVRVAWLFLFSKFIELL 127
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTV +VR Q++FLH++H
Sbjct 128 DTVLFVVRKKNGQITFLHIFH 148
> dre:334217 elovl7a, MGC174838, elovl7, fi36f02, wu:fi36f02,
zgc:55879; ELOVL family member 7, elongation of long chain fatty
acids (yeast) a; K10250 elongation of very long chain fatty
acids protein 7
Length=288
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 44/81 (54%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
+YNI V L ++ Y+F C+ + + RMA V W +Y K++ +L
Sbjct 70 VYNIFVVSLSVYMCYEFLMAGWGTGYTFGCDLVDYSQSPKAMRMASVCWLYYFSKFIVML 129
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF ++R +Q++FLHV+H
Sbjct 130 DTVFFVLRKKPKQITFLHVFH 150
> dre:767653 MGC153394; zgc:153394
Length=268
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 43/82 (52%), Gaps = 6/82 (7%)
Query 26 IYNIAQVILCGFLVAR-VFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDL 80
IYN + V L ++ W A YS +C + RMA V W+FY K ++L
Sbjct 71 IYNFSMVCLSAYMFYEFTVSSWLAS-YSLLCQPVDYTENPLPMRMARVCWWFYFSKVIEL 129
Query 81 LDTVFMIVRGNWRQVSFLHVYH 102
DT+F I+R Q++FLHVYH
Sbjct 130 ADTMFFILRKKNNQLTFLHVYH 151
> dre:554145 im:7139490; zgc:112263; K10249 elongation of very
long chain fatty acids protein 4
Length=264
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 50/96 (52%), Gaps = 6/96 (6%)
Query 13 LLEKMGPVFV--CQAIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMA 66
L++ M PV + +YN + V L ++ YS++C + + RMA
Sbjct 52 LMKNMEPVNLKGLLIVYNFSMVGLSVYMFHEFLVTSWLANYSYLCQPVDYSTSPLGMRMA 111
Query 67 EVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
V W+F+ K ++L DTVF I+R Q++FLHVYH
Sbjct 112 NVCWWFFFSKVIELSDTVFFILRKKNSQLTFLHVYH 147
> xla:379510 hypothetical protein MGC64517; K10247 elongation
of very long chain fatty acids protein 1
Length=290
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 44/81 (54%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
+YN + V L ++V Y++ C+ +V + RM +V W F K+++LL
Sbjct 68 VYNFSLVALSAYIVYEFLMSGWLTGYTWRCDPVDVSDKPMALRMVQVAWLFLFSKFIELL 127
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTVF +VR Q++FLH+ H
Sbjct 128 DTVFFVVRKKNSQITFLHIIH 148
> dre:678614 elovl2, MGC136352, MGC158526, zgc:136352; elongation
of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
2; K10205 elongation of very long chain fatty acids
protein 2 [EC:2.3.1.-]
Length=260
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 45/80 (56%), Gaps = 3/80 (3%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNV---PNTRMAEVTWFFYMLKYVDLLD 82
+YN + +L +++ + + Y C + + R+A+V W++Y K ++ LD
Sbjct 70 LYNFSVTVLSFYMLVELISAVWSAGYRLQCQALDEVGEADIRVAKVLWWYYFSKLIEFLD 129
Query 83 TVFMIVRGNWRQVSFLHVYH 102
T+F+++R Q+SFLHVYH
Sbjct 130 TIFIVLRKKNSQISFLHVYH 149
> xla:447388 elovl2, MGC84669; elongation of very long chain fatty
acids (FEN1/Elo2, SUR4/Elo3)-like 2; K10205 elongation
of very long chain fatty acids protein 2 [EC:2.3.1.-]
Length=296
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 52/98 (53%), Gaps = 9/98 (9%)
Query 9 REKPLLEKMGPVFVCQAIYNIAQVILCGF-LVARVFDVWRAEEYSFICNRFNVP---NTR 64
+++P G + V YN+ +L + L+ + W+ Y+ C + + R
Sbjct 57 KDRPAFSLRGHLIV----YNLGVTLLSFYMLIELILSTWQGA-YNLQCQNLDSAGEADVR 111
Query 65 MAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+A+V W++Y K ++ +DT+F ++R Q++FLHVYH
Sbjct 112 VAKVLWWYYFSKAIEFMDTIFFVLRKKNSQITFLHVYH 149
> dre:393425 elovl5, MGC63549, zgc:63549; ELOVL family member
5, elongation of long chain fatty acids (yeast); K10244 elongation
of very long chain fatty acids protein 5 [EC:2.3.1.-]
Length=291
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 13/103 (12%)
Query 13 LLEKMGP-------VFVCQAI---YNIAQVILCGFLVARVFDVWRAEEYSFICNRFNV-- 60
L+ MGP + C+A+ YN+ +L ++ + Y+F C +
Sbjct 44 LIVWMGPKYMKNRQAYSCRALLVPYNLCLTLLSLYMFYELVMSVYQGGYNFFCQNTHSGG 103
Query 61 -PNTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+ RM V W++Y K ++ +DT F I+R N Q++FLHVYH
Sbjct 104 DADNRMMNVLWWYYFSKLIEFMDTFFFILRKNNHQITFLHVYH 146
> hsa:79993 ELOVL7, FLJ23563; ELOVL family member 7, elongation
of long chain fatty acids (yeast); K10250 elongation of very
long chain fatty acids protein 7
Length=281
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 35/56 (62%), Gaps = 4/56 (7%)
Query 51 YSFICNRFNVPNT----RMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
YSF C+ + + RMA W +Y K+++LLDT+F ++R QV+FLHV+H
Sbjct 95 YSFRCDIVDYSRSPTALRMARTCWLYYFSKFIELLDTIFFVLRKKNSQVTFLHVFH 150
> hsa:64834 ELOVL1, Ssc1; elongation of very long chain fatty
acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1; K10247 elongation
of very long chain fatty acids protein 1
Length=279
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 4/81 (4%)
Query 26 IYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLL 81
+YN + V L ++V Y++ C+ + N+ RM V W F K+++L+
Sbjct 64 VYNFSLVALSLYIVYEFLMSGWLSTYTWRCDPVDYSNSPEALRMVRVAWLFLFSKFIELM 123
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DTV I+R QV+FLHV+H
Sbjct 124 DTVIFILRKKDGQVTFLHVFH 144
> mmu:74559 Elovl7, 9130013K24Rik, AI840082; ELOVL family member
7, elongation of long chain fatty acids (yeast); K10250 elongation
of very long chain fatty acids protein 7
Length=281
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 48/96 (50%), Gaps = 6/96 (6%)
Query 13 LLEKMGPVFVCQAI--YNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNT----RMA 66
L+E P + +A+ YN V+ ++ YSF C+ + + RM
Sbjct 55 LMENRKPFELKKAMITYNFFIVLFSVYMCYEFVMSGWGTGYSFRCDIVDYSQSPRAMRMV 114
Query 67 EVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
W +Y K+++LLDT+F ++R QV+FLHV+H
Sbjct 115 HTCWLYYFSKFIELLDTIFFVLRKKNSQVTFLHVFH 150
> xla:734950 elovl5, MGC131143; ELOVL family member 5, elongation
of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like); K10244
elongation of very long chain fatty acids protein 5 [EC:2.3.1.-]
Length=295
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 52/101 (51%), Gaps = 9/101 (8%)
Query 9 REKPLLEKMGPVFVCQAI---YNIAQVILCGFLVAR-VFDVWRAEEYSFICNRFNV---P 61
R ++ PV C++I YN+ +L ++ V VW Y+F C +
Sbjct 48 RGPKYMQNRQPV-SCRSILVVYNLGLTLLSFYMFYELVTGVWEGG-YNFFCQDTHSGGDA 105
Query 62 NTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+T++ V W++Y K ++ +DT F I+R N Q++ LHVYH
Sbjct 106 DTKIIRVLWWYYFSKLIEFMDTFFFILRKNNHQITVLHVYH 146
> hsa:60481 ELOVL5, HELO1, dJ483K16.1; ELOVL family member 5,
elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like,
yeast); K10244 elongation of very long chain fatty acids
protein 5 [EC:2.3.1.-]
Length=299
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 46/89 (51%), Gaps = 8/89 (8%)
Query 21 FVCQAI---YNIAQVILCGFLVAR-VFDVWRAEEYSFICNRFNV---PNTRMAEVTWFFY 73
F C+ I YN+ +L ++ V VW + Y+F C + ++ V W++Y
Sbjct 59 FSCRGILVVYNLGLTLLSLYMFCELVTGVWEGK-YNFFCQGTRTAGESDMKIIRVLWWYY 117
Query 74 MLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
K ++ +DT F I+R N Q++ LHVYH
Sbjct 118 FSKLIEFMDTFFFILRKNNHQITVLHVYH 146
> mmu:68801 Elovl5, 1110059L23Rik, AI747313, AU043003, HELO1;
ELOVL family member 5, elongation of long chain fatty acids
(yeast); K10244 elongation of very long chain fatty acids protein
5 [EC:2.3.1.-]
Length=299
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 46/89 (51%), Gaps = 8/89 (8%)
Query 21 FVCQAI---YNIAQVILCGFLVAR-VFDVWRAEEYSFICN---RFNVPNTRMAEVTWFFY 73
F C+ I YN+ +L ++ V VW + Y+F C + ++ V W++Y
Sbjct 59 FSCRGILQLYNLGLTLLSLYMFYELVTGVWEGK-YNFFCQGTRSAGESDMKIIRVLWWYY 117
Query 74 MLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
K ++ +DT F I+R N Q++ LHVYH
Sbjct 118 FSKLIEFMDTFFFILRKNNHQITVLHVYH 146
> xla:734710 hypothetical protein MGC114802; K10244 elongation
of very long chain fatty acids protein 5 [EC:2.3.1.-]
Length=295
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 44/81 (54%), Gaps = 5/81 (6%)
Query 26 IYNIAQVILCGFLVAR-VFDVWRAEEYSFICNRFNV---PNTRMAEVTWFFYMLKYVDLL 81
+YN+ +L ++ V VW Y+F C + +T++ V W++Y K ++ +
Sbjct 67 VYNLVLTLLSLYMFYELVTGVWEGG-YNFFCQDTHSGGDADTKIIRVLWWYYFSKLIEFM 125
Query 82 DTVFMIVRGNWRQVSFLHVYH 102
DT F I+R N Q++ LHVYH
Sbjct 126 DTFFFILRKNNHQMTVLHVYH 146
> mmu:54325 Elovl1, AA407424, BB151133, Ssc1; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
1; K10247 elongation of very long chain fatty acids protein
1
Length=202
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 34/58 (58%), Gaps = 4/58 (6%)
Query 49 EEYSFICNRFNVPNT----RMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
Y++ C+ + N+ RM V W F + K ++L+DTV I+R QV+FLHV+H
Sbjct 10 STYTWRCDPIDFSNSPEALRMVRVAWLFMLSKVIELMDTVIFILRKKDGQVTFLHVFH 67
> sce:YJL196C ELO1; Elo1p; K10245 fatty acid elongase 2 [EC:2.3.1.-]
Length=310
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 7/60 (11%)
Query 45 VWRAEEYSFICN--RFNVPNTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
V+R Y +CN + P M + + YM K+V+ DTV M+++ R+++FLH YH
Sbjct 121 VYRHGLYFAVCNVESWTQP---METLYYLNYMTKFVEFADTVLMVLKH--RKLTFLHTYH 175
> sce:YLR372W SUR4, APA1, ELO3, SRE1, VBM1; Elongase, involved
in fatty acid and sphingolipid biosynthesis; synthesizes very
long chain 20-26-carbon fatty acids from C18-CoA primers;
involved in regulation of sphingolipid biosynthesis; K10246
fatty acid elongase 3 [EC:2.3.1.-]
Length=345
Score = 33.5 bits (75), Expect = 0.16, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 28/39 (71%), Gaps = 2/39 (5%)
Query 64 RMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
++ + + Y+ K+V+L+DTVF+++R +++ FLH YH
Sbjct 152 KLVTLYYLNYLTKFVELIDTVFLVLRR--KKLLFLHTYH 188
> ath:AT3G06460 GNS1/SUR4 membrane family protein
Length=298
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 72 FYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
FY+ K ++ +DT+ +I+ + +++SFLHVYH
Sbjct 124 FYLSKILEFVDTLLIILNKSIQRLSFLHVYH 154
> ath:AT2G04650 ADP-glucose pyrophosphorylase family protein
Length=406
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 29/63 (46%), Gaps = 10/63 (15%)
Query 11 KPLLEKMG------PVFVCQAIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNTR 64
KPL+ G P+ C+ I N+AQ+ L GF R F ++ S I N +P
Sbjct 30 KPLIPLAGQPMIHHPISACKKISNLAQIFLIGFYEEREFALY----VSSISNELKIPVRY 85
Query 65 MAE 67
+ E
Sbjct 86 LKE 88
> ath:AT3G06470 GNS1/SUR4 membrane family protein
Length=278
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 72 FYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
FY+ K ++ DT+ +I+ + +++SFLHVYH
Sbjct 127 FYLSKILEFGDTILIILGKSIQRLSFLHVYH 157
> cel:D2024.3 elo-3; fatty acid ELOngation family member (elo-3)
Length=320
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 48/91 (52%), Gaps = 14/91 (15%)
Query 19 PVFVCQAIYNIAQVILCGFL--VARVFDVWRAEEYSFICNRFNVPNTRMAE-VTWF---- 71
P+FV + I ++ GFL W AE SF ++++ ++ A+ VT F
Sbjct 70 PLFVWNSFLAIFSIL--GFLRMTPEFVWSWSAEGNSF---KYSICHSSYAQGVTGFWTEQ 124
Query 72 FYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
F M K +L+DT+F+++R R + FLH YH
Sbjct 125 FAMSKLFELIDTIFIVLRK--RPLIFLHWYH 153
> hsa:83401 ELOVL3, CIG-30, CIG30, MGC21435; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
3; K10248 elongation of very long chain fatty acids protein
3
Length=270
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 47/97 (48%), Gaps = 7/97 (7%)
Query 9 REKPLLEKMGPVFV---CQAIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVPNTRM 65
+E+ GP+ + C AI++I + ++ V ++ +C + N+ +
Sbjct 56 KERKGFNLQGPLILWSFCLAIFSILGAVRMWGIMGTVLLTGGLKQT--VCFINFIDNSTV 113
Query 66 AEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
+W F + K ++L DT F+I+R R + F+H YH
Sbjct 114 KFWSWVFLLSKVIELGDTAFIILRK--RPLIFIHWYH 148
> tgo:TGME49_005350 fatty acid elongation protein, putative
Length=318
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 25/39 (64%), Gaps = 4/39 (10%)
Query 66 AEVTWFFYML--KYVDLLDTVFMIVRGNWRQVSFLHVYH 102
A W F + KY +LLDT+F+++R R ++FLH YH
Sbjct 141 ASGLWIFLFIYSKYFELLDTLFIVLRK--RPLNFLHWYH 177
> ath:AT1G74910 ADP-glucose pyrophosphorylase family protein
Length=415
Score = 31.2 bits (69), Expect = 0.85, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 26/57 (45%), Gaps = 10/57 (17%)
Query 11 KPLLEKMG------PVFVCQAIYNIAQVILCGFLVARVFDVWRAEEYSFICNRFNVP 61
KPL G P+ C+ I N+AQ+ L GF R F ++ S I N VP
Sbjct 33 KPLFPIAGQPMVHHPISACKRIPNLAQIYLVGFYEEREFALY----VSAISNELKVP 85
> cel:F56H11.4 elo-1; fatty acid ELOngation family member (elo-1)
Length=288
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 2/48 (4%)
Query 55 CNRFNVPNTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
C F+ W F K +L+DT+F+++R R + FLH YH
Sbjct 113 CKVFDFTKGENGYWVWLFMASKLFELVDTIFLVLRK--RPLMFLHWYH 158
> cpv:cgd8_4630 7 pass integral membrane proteinwith FLHWFHH motif
shared with fatty-acyl elongase
Length=323
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 4/53 (7%)
Query 52 SFICNRFNVPNTRMAEVTWF--FYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
S IC+ P T+ W F KY++L+DT F+I R + +SFLH +H
Sbjct 127 SSICSPPIAPLTKGPAGLWLSLFIYSKYIELIDTFFIIARK--KSLSFLHWFH 177
> pfa:PFL0295c conserved Plasmodium membrane protein
Length=1082
Score = 31.2 bits (69), Expect = 0.92, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 29/51 (56%), Gaps = 4/51 (7%)
Query 48 AEEYSFICNR----FNVPNTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQ 94
++E++ +R + + +T A +TW ++KY LD V++++R +Q
Sbjct 891 SQEFAITASRIWLQYTIASTLSAALTWPLALIKYASNLDNVYLLIRERAQQ 941
> sce:YCR034W FEN1, ELO2, GNS1, VBM2; Fatty acid elongase, involved
in sphingolipid biosynthesis; acts on fatty acids of up
to 24 carbons in length; mutations have regulatory effects
on 1,3-beta-glucan synthase, vacuolar ATPase, and the secretory
pathway; K10245 fatty acid elongase 2 [EC:2.3.1.-]
Length=347
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 10/30 (33%), Positives = 24/30 (80%), Gaps = 2/30 (6%)
Query 73 YMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
Y++K+++ +DT F++++ ++++FLH YH
Sbjct 154 YIVKFIEFIDTFFLVLKH--KKLTFLHTYH 181
> pfa:PFA_0455c fatty acid elongation protein, GNS1/SUR4 family,
putative
Length=322
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 25/32 (78%), Gaps = 2/32 (6%)
Query 71 FFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
FF + KY +L+DT+F+I++ ++++FLH +H
Sbjct 150 FFIISKYFELIDTLFLILKK--KEITFLHWFH 179
> cel:F41H10.8 elo-6; fatty acid ELOngation family member (elo-6)
Length=274
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Query 62 NTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
N TW F + K + DT+F+I+R + + FLH YH
Sbjct 106 NGLSGMFTWLFVLSKVAEFGDTLFIILRK--KPLMFLHWYH 144
> cel:C40H1.4 elo-4; fatty acid ELOngation family member (elo-4)
Length=291
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query 51 YSFICNRFNVPNTRMAEVTWFFYMLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
Y +C N P A ++ F + K V+L DT+F+I+R R + FLH YH
Sbjct 117 YKTLCYSCN-PTDVAAFWSFAFALSKIVELGDTMFIILRK--RPLIFLHYYH 165
> cel:F11E6.5 elo-2; fatty acid ELOngation family member (elo-2)
Length=274
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 8/84 (9%)
Query 25 AIYNIAQVILCGF----LVARVFDVWRAEEY--SFICNRFNVPNTRMAEVTWFFYMLKYV 78
A++N + G L+ +F V+ + + S+ N + W F M K
Sbjct 67 ALWNFGFSLFSGIAAYKLIPELFGVFMKDGFVASYCQNENYYTDASTGFWGWAFVMSKAP 126
Query 79 DLLDTVFMIVRGNWRQVSFLHVYH 102
+L DT+F+++R + V F+H YH
Sbjct 127 ELGDTMFLVLRK--KPVIFMHWYH 148
> cel:F41H10.7 elo-5; fatty acid ELOngation family member (elo-5)
Length=286
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 45/98 (45%), Gaps = 8/98 (8%)
Query 9 REKPLLEKMGPVFVCQAIYNIAQVILCGFLVA--RVFDVWRAEEYSFICNRFNVPNTRMA 66
+++ + P+ + I ++ L GFL + V R + +S + + T
Sbjct 57 KDRKAFDLSTPLNIWNGI--LSTFSLLGFLFTFPTLLSVIRKDGFSHTYSHVSELYTDST 114
Query 67 EVTWFFY--MLKYVDLLDTVFMIVRGNWRQVSFLHVYH 102
W F + K +LLDTVF+++R R + F+H YH
Sbjct 115 SGYWIFLWVISKIPELLDTVFIVLRK--RPLIFMHWYH 150
Lambda K H
0.333 0.142 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40