bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0914_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
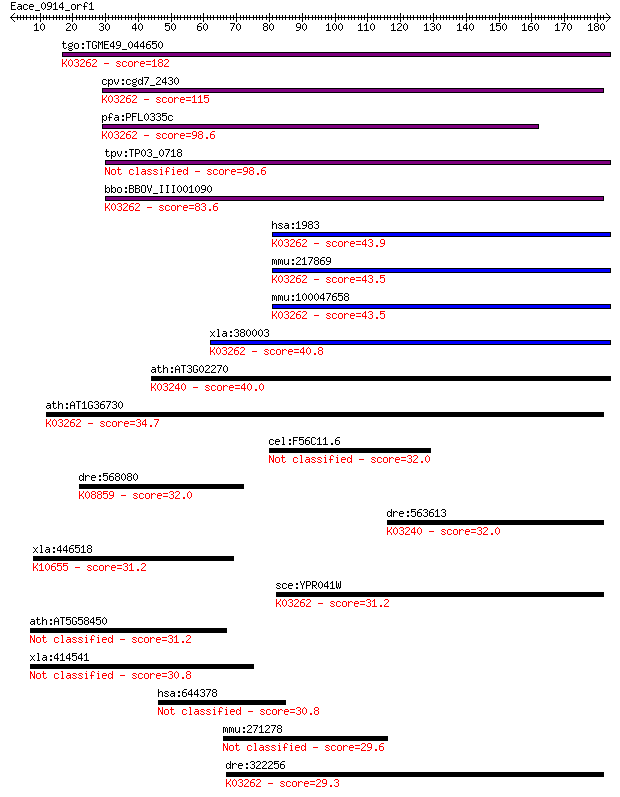
tgo:TGME49_044650 eukaryotic translation initiation factor 5, ... 182 5e-46
cpv:cgd7_2430 translation initiation factor eIF-5; Tif5p, ZnR+... 115 9e-26
pfa:PFL0335c eukaryotic translation initiation factor 5, putat... 98.6 1e-20
tpv:TP03_0718 eukaryotic translation initiation factor 5 98.6
bbo:BBOV_III001090 17.m07123; eukaryotic translation initiatio... 83.6 4e-16
hsa:1983 EIF5, EIF-5A; eukaryotic translation initiation facto... 43.9 3e-04
mmu:217869 Eif5, 2810011H21Rik, D12Ertd549e, MGC36374, MGC3650... 43.5 4e-04
mmu:100047658 eukaryotic translation initiation factor 5-like;... 43.5 4e-04
xla:380003 eif5, MGC52788; eukaryotic translation initiation f... 40.8 0.002
ath:AT3G02270 eIF4-gamma/eIF5/eIF2-epsilon domain-containing p... 40.0 0.005
ath:AT1G36730 eukaryotic translation initiation factor 5, puta... 34.7 0.20
cel:F56C11.6 hypothetical protein 32.0 1.2
dre:568080 stk35, MGC153287, im:6905214, zgc:153287; serine/th... 32.0 1.2
dre:563613 eif2b5, fd14g07, wu:fd14g07; eukaryotic translation... 32.0 1.2
xla:446518 trim63, MGC80210, rnf30; tripartite motif containin... 31.2 2.0
sce:YPR041W TIF5, SUI5; Tif5p; K03262 translation initiation f... 31.2 2.1
ath:AT5G58450 binding 31.2 2.3
xla:414541 pdik1l-b, MGC81117, pdik1-b, pdik1l; PDLIM1 interac... 30.8 2.7
hsa:644378 GCNT6, bA421M1.3; glucosaminyl (N-acetyl) transfera... 30.8 2.8
mmu:271278 6230424I18Rik, MGC37978; cDNA sequence BC024139 29.6 6.6
dre:322256 eif5, fb37c12, fb54h04, wu:fb37c12, wu:fb54h04, zgc... 29.3 8.8
> tgo:TGME49_044650 eukaryotic translation initiation factor 5,
putative ; K03262 translation initiation factor 5
Length=414
Score = 182 bits (462), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 88/167 (52%), Positives = 116/167 (69%), Gaps = 0/167 (0%)
Query 17 SGSVSEGPKLVVKEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVELRMLQVSQDFDA 76
S ++ KL+ KEAL PE+ +V RLRN+ + +V AFF E+RMLQVSQDFD+
Sbjct 239 SSALDSEKKLMHKEALVFDSPELKDVIERLRNVVTRSETVDLAAFFHEMRMLQVSQDFDS 298
Query 77 KCRLFVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGF 136
KCR++V L+A+F + +TP L+ +MP+++K D+SV +SD++ A +C E T MT F
Sbjct 299 KCRIYVTLAAIFNDDMTPTSLEARMPYILKVLDTSVSASDVLDAFGFYCQEKGGTAMTSF 358
Query 137 PYLLQRMYNAELLEAEDILNYYNADTTDPVTLKCKTFAEPFLQWLAE 183
PY LQ++YNAE LEAEDIL YY AD DPV CK AEPFLQWLAE
Sbjct 359 PYCLQKLYNAEALEAEDILKYYAADKEDPVFSACKKQAEPFLQWLAE 405
> cpv:cgd7_2430 translation initiation factor eIF-5; Tif5p, ZnR+W2
domains ; K03262 translation initiation factor 5
Length=460
Score = 115 bits (288), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 62/164 (37%), Positives = 93/164 (56%), Gaps = 11/164 (6%)
Query 29 KEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVLSALF 88
K+ LT PEI +V RL N V+P+ FF ELR+LQVSQDFD R V+L+ LF
Sbjct 209 KDVLTFESPEIVDVIERLTNFMQKNEEVSPEDFFSELRVLQVSQDFDEFMRFSVLLAVLF 268
Query 89 --KEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQRMYNA 146
K+ + P+ ++P++ K + S+RS+ II E++C E + + +PYL+Q++Y+A
Sbjct 269 GLKDPIDPKVFKLRIPYISKAVEGSLRSNAIISIFESYCVEKNPSTLATYPYLIQQLYDA 328
Query 147 ELLEAEDILNYYNAD---------TTDPVTLKCKTFAEPFLQWL 181
++L + IL YY D +T K K EPF+ WL
Sbjct 329 DILSEKVILKYYQKDAPTSWVSGCSTQDTFEKAKKSCEPFVDWL 372
> pfa:PFL0335c eukaryotic translation initiation factor 5, putative;
K03262 translation initiation factor 5
Length=565
Score = 98.6 bits (244), Expect = 1e-20, Method: Composition-based stats.
Identities = 49/133 (36%), Positives = 84/133 (63%), Gaps = 0/133 (0%)
Query 29 KEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVLSALF 88
KE L PEI EV R++ +F + + + ELR+LQVSQ FD+KCR+F+ L +LF
Sbjct 253 KECLHFGSPEIKEVIERMKTIFKESTQMNDIQYAEELRVLQVSQCFDSKCRVFICLCSLF 312
Query 89 KEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQRMYNAEL 148
++ ++ E L++ + ++ K D+SV + DI ALE + + ++ +PY+LQ +YN ++
Sbjct 313 EDKISKELLEKNIKYIKKINDTSVTTMDIFLALEYYVNKVAINSLSIYPYILQVLYNNDI 372
Query 149 LEAEDILNYYNAD 161
E++DI+ Y+ D
Sbjct 373 FESKDIIKRYDDD 385
> tpv:TP03_0718 eukaryotic translation initiation factor 5
Length=385
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 56/161 (34%), Positives = 87/161 (54%), Gaps = 8/161 (4%)
Query 30 EALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVLSALFK 89
E L++ PE++E++ RLR F+ T + F EL MLQ+SQ FD R+F+ LSA+F
Sbjct 224 EDLSVESPELSEIATRLR-CFLEKGDRTCEDFSEELHMLQISQGFDNVSRMFIALSAIFN 282
Query 90 EG--LTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQRMYNAE 147
LT E ++ V S+ + I ALE F F+S ++T +PY Q +Y+ +
Sbjct 283 SDVPLTLELFKDRIQLVKAATGYSMTPKEKITALEYFIFQSNPQEITNYPYYSQLLYSHD 342
Query 148 LLEAEDILNYYN-----ADTTDPVTLKCKTFAEPFLQWLAE 183
+LE D + YYN + + + L+ K PF++WL E
Sbjct 343 ILEGSDFIRYYNKSNRHSGNLNNIFLRAKETIAPFIEWLQE 383
> bbo:BBOV_III001090 17.m07123; eukaryotic translation initiation
factor 5; K03262 translation initiation factor 5
Length=379
Score = 83.6 bits (205), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 47/157 (29%), Positives = 80/157 (50%), Gaps = 5/157 (3%)
Query 30 EALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVLSALFK 89
E LTI PEI + R+ + P F EL MLQ+SQDFD C+++V L A+F
Sbjct 210 EKLTIESPEIQDAIERITSSLKGGKFQQPAEFAEELHMLQLSQDFDNICKVYVALCAIFS 269
Query 90 EGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQRMYNAELL 149
+ + L+ K+ ++ + V+ D++ ++ F + Q+ +PY+ + +Y ++L
Sbjct 270 DKFDVQELEDKINLLIPLLEYGVQPKDVLTSMAVFVVKLYPQQLDDYPYICRALYASDLA 329
Query 150 EAEDILNYYNADTTDPVTL-----KCKTFAEPFLQWL 181
+ DI +Y T +L + K AEPF+ WL
Sbjct 330 DGGDICRFYAKSTRFDGSLAEAYIRTKEKAEPFVAWL 366
> hsa:1983 EIF5, EIF-5A; eukaryotic translation initiation factor
5; K03262 translation initiation factor 5
Length=431
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Query 81 FVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLL 140
V+ LF E + + + F+ C ++ +++ LE + ++ P++L
Sbjct 278 LVLTEVLFNEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLECVVAMHQAQLISKIPHIL 337
Query 141 QRMYNAELLEAEDILNYYNADTTDPVT----LKCKTFAEPFLQWLAE 183
+ MY+A+LLE E I+++ + V+ + + AEPF++WL E
Sbjct 338 KEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 384
> mmu:217869 Eif5, 2810011H21Rik, D12Ertd549e, MGC36374, MGC36509;
eukaryotic translation initiation factor 5; K03262 translation
initiation factor 5
Length=429
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Query 81 FVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLL 140
V+ LF E + + + F+ C ++ +++ LE + ++ P++L
Sbjct 276 LVLTEVLFDEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLECVVAMHQAQLISKIPHIL 335
Query 141 QRMYNAELLEAEDILNYYNADTTDPVT----LKCKTFAEPFLQWLAE 183
+ MY+A+LLE E I+++ + V+ + + AEPF++WL E
Sbjct 336 KEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 382
> mmu:100047658 eukaryotic translation initiation factor 5-like;
K03262 translation initiation factor 5
Length=429
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 53/107 (49%), Gaps = 4/107 (3%)
Query 81 FVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLL 140
V+ LF E + + + F+ C ++ +++ LE + ++ P++L
Sbjct 276 LVLTEVLFDEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLECVVAMHQAQLISKIPHIL 335
Query 141 QRMYNAELLEAEDILNYYNADTTDPVT----LKCKTFAEPFLQWLAE 183
+ MY+A+LLE E I+++ + V+ + + AEPF++WL E
Sbjct 336 KEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 382
> xla:380003 eif5, MGC52788; eukaryotic translation initiation
factor 5; K03262 translation initiation factor 5
Length=434
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/127 (22%), Positives = 62/127 (48%), Gaps = 5/127 (3%)
Query 62 FVELRMLQVSQDFDAKCRLFVVLS-ALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYA 120
F + +L ++ D K +VLS LF + + + + F+ C ++ +++
Sbjct 254 FCDKDILAEAERLDVKAMGPLVLSEVLFDDKIRDQIKKYRRHFLRFCHNNKKAQRYLLHG 313
Query 121 LENFCFESEETQMTGFPYLLQRMYNAELLEAEDILNYYNADTTDPVTLK----CKTFAEP 176
E + ++ P++L+ MY+A+LLE E I+++ + V+ + + A+P
Sbjct 314 FECVIDMHQSHLLSKIPHILKEMYDADLLEEEVIISWAEKPSKKYVSKELAKDIRAKADP 373
Query 177 FLQWLAE 183
F++WL E
Sbjct 374 FIKWLKE 380
> ath:AT3G02270 eIF4-gamma/eIF5/eIF2-epsilon domain-containing
protein; K03240 translation initiation factor eIF-2B subunit
epsilon
Length=676
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 45/147 (30%), Positives = 65/147 (44%), Gaps = 20/147 (13%)
Query 44 NRLRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVLSALFKEGLTPEGLDQKMPF 103
N LR + + A F + L VS + L+ S++ +GL + F
Sbjct 536 NSLRLSYNMESAHCAGAIFYSMMKLAVSTPHSSINDLYRNASSIITRW---KGL---LGF 589
Query 104 VVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQRMYNAE--LLEAEDILNYYN-- 159
VK D + ++I LE C ES T F ++L+ MY E LL+ IL + +
Sbjct 590 YVKKSDEQI---EVISRLEEMCEESAHELGTLFAHILRYMYEEENDLLQEVAILRWSDEK 646
Query 160 --ADTTDPVTLK-CKTFAEPFLQWLAE 183
AD +D V LK C EPF+ WL E
Sbjct 647 AGADESDKVYLKQC----EPFITWLKE 669
> ath:AT1G36730 eukaryotic translation initiation factor 5, putative
/ eIF-5, putative; K03262 translation initiation factor
5
Length=439
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 37/179 (20%), Positives = 81/179 (45%), Gaps = 19/179 (10%)
Query 12 KEKKQSGSVSEGPKLVVKEALTIRCPEIAE--VSNRLRNMFITTPSVTPDAFFVELRMLQ 69
+EKK V + P+ V E + PE A + N ++ + + S T +L+
Sbjct 261 EEKKPVAEVKKAPEQV-HENGNSKIPENAHEKLVNEIKELLSSGSSPT------QLKTAL 313
Query 70 VSQDFDAKCRLFVVLSALFK---EGLTPEGLDQK---MPFVVKCCDSSVRSS-DIIYALE 122
S + + ++ + SALF +G E + +K + ++ ++ + ++ +E
Sbjct 314 ASNSANPQEKMDALFSALFGGTGKGFAKEVIKKKKYLLALMMMQEEAGAPAQMGLLNGIE 373
Query 123 NFCFESEETQMTGFPYLLQRMYNAELLEAEDILNYYNADTTDPVTLKCKTFAEPFLQWL 181
+FC ++ +++ +Y+ ++L+ + I+ +YN LK T PF++WL
Sbjct 374 SFCMKASAEAAKEVALVIKGLYDEDILDEDVIVEWYNKGVKSSPVLKNVT---PFIEWL 429
> cel:F56C11.6 hypothetical protein
Length=548
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 80 LFVVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFES 128
F+ L AL K PEG+ + M + K CD R+ D++ + +F F+
Sbjct 338 FFIALGALSK---NPEGIKKFMGRIFKECDYGERADDVLQMVYDFYFKG 383
> dre:568080 stk35, MGC153287, im:6905214, zgc:153287; serine/threonine
kinase 35 (EC:2.7.11.1); K08859 serine/threonine kinase
35 [EC:2.7.11.1]
Length=406
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 22 EGPKLVVKEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVELRMLQVS 71
E PK+V+ R V L++M P PDAF +E+RM QV+
Sbjct 354 ENPKMVLHIPQKTRSIMSEGVKRLLQDMLAVNPQDRPDAFQLEVRMDQVT 403
> dre:563613 eif2b5, fd14g07, wu:fd14g07; eukaryotic translation
initiation factor 2B, subunit 5 epsilon; K03240 translation
initiation factor eIF-2B subunit epsilon
Length=703
Score = 32.0 bits (71), Expect = 1.2, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 34/68 (50%), Gaps = 3/68 (4%)
Query 116 DIIYALENFCFESEETQMTGFPYLLQRMYNAELLEAEDILNYYNADTTDPVTLKCKTFAE 175
D + ++E F ++T +L MY E+LE + I+ ++ +T + K +T A
Sbjct 627 DCLTSMEE-VFLEQDTHWAALVKVLMNMYQLEILEEDVIMRWFTQSSTTDKSQKLRTNAG 685
Query 176 --PFLQWL 181
F+QWL
Sbjct 686 LLKFIQWL 693
> xla:446518 trim63, MGC80210, rnf30; tripartite motif containing
63; K10655 tripartite motif-containing protein 63 [EC:6.3.2.19]
Length=356
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 1/62 (1%)
Query 8 EKKKKEKKQSGSVSEGPKLVVKEALTIRCPE-IAEVSNRLRNMFITTPSVTPDAFFVELR 66
E+KK Q S +E KL + +L +RC + + SN L+N F P F +E +
Sbjct 218 EEKKTHLVQQISQAEEEKLNIVRSLIVRCQQQLQSSSNELQNAFQAMEQTGPANFLLESK 277
Query 67 ML 68
L
Sbjct 278 QL 279
> sce:YPR041W TIF5, SUI5; Tif5p; K03262 translation initiation
factor 5
Length=405
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/104 (21%), Positives = 46/104 (44%), Gaps = 5/104 (4%)
Query 82 VVLSALFKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFCFESEETQMTGFPYLLQ 141
V+ LF E + E + + F K + + + +E F + + P +L
Sbjct 290 VLAQCLFDEDIVNE-IAEHNAFFTKILVTPEYEKNFMGGIERFLGLEHKDLIPLLPKILV 348
Query 142 RMYNAELLEAEDILNYYNADTT----DPVTLKCKTFAEPFLQWL 181
++YN +++ E+I+ + + V+ K + A+PF+ WL
Sbjct 349 QLYNNDIISEEEIMRFGTKSSKKFVPKEVSKKVRRAAKPFITWL 392
> ath:AT5G58450 binding
Length=1065
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Query 7 SEKKKKEKKQSGSVSEGP-KLVVKEALTIRCPEIAEVSNRLRNMFITTPSVTPDAFFVEL 65
S KKKK+ + S +S P +K+++ + C I +VSN L N + F L
Sbjct 926 SGKKKKKNQHSDQLSSSPISQAIKDSIQLLCSTIQDVSNWLLNQLNNPEDGQVEGFLTTL 985
Query 66 R 66
+
Sbjct 986 K 986
> xla:414541 pdik1l-b, MGC81117, pdik1-b, pdik1l; PDLIM1 interacting
kinase 1 like (EC:2.7.11.1)
Length=339
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 40/81 (49%), Gaps = 13/81 (16%)
Query 7 SEKKKKEKKQSGSVSEGPKLV-VKEALT------IRCPEIAEVSNR-----LRNMFITTP 54
++ K++ G V G ++V V EAL + P + NR LR M P
Sbjct 255 TDTHTKKRLLGGYVQRGAQVVPVGEALLENPKLELLIPVKKKSMNRRMKQLLRQMLSANP 314
Query 55 SVTPDAFFVELRMLQVS-QDF 74
PDAF +EL+++Q++ +DF
Sbjct 315 QERPDAFQLELKLIQIAFRDF 335
> hsa:644378 GCNT6, bA421M1.3; glucosaminyl (N-acetyl) transferase
6
Length=391
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 46 LRNMFITTPSVTPDAFFVELRMLQVSQDFDAKCRLFVVL 84
L N +ITTP T +A F ++ +SQDFD LF +
Sbjct 81 LENHYITTPLSTEEAAFPLAYVMTISQDFDTFEWLFWAI 119
> mmu:271278 6230424I18Rik, MGC37978; cDNA sequence BC024139
Length=772
Score = 29.6 bits (65), Expect = 6.6, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 33/55 (60%), Gaps = 5/55 (9%)
Query 66 RMLQVSQDFDAKCR-LFVVLSA----LFKEGLTPEGLDQKMPFVVKCCDSSVRSS 115
+++ +Q F+ C+ L V LS+ L + L P+GL++ +P++ + C+ V S+
Sbjct 178 QLVPAAQSFETACKALLVRLSSSEQLLAELWLGPKGLEESLPYLQEVCEGVVAST 232
> dre:322256 eif5, fb37c12, fb54h04, wu:fb37c12, wu:fb54h04, zgc:56606,
zgc:77026; eukaryotic translation initiation factor
5; K03262 translation initiation factor 5
Length=429
Score = 29.3 bits (64), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 26/120 (21%), Positives = 53/120 (44%), Gaps = 5/120 (4%)
Query 67 MLQVSQDFDAKCRLFVVLSAL-FKEGLTPEGLDQKMPFVVKCCDSSVRSSDIIYALENFC 125
+L ++ D K ++LS L F E + + K F+ C ++ ++ E
Sbjct 256 ILAEAERLDVKAMGPLILSELLFDENIRDQIKKYKRHFLRFCHNNKKAQKYLLGGFECLV 315
Query 126 FESEETQMTGFPYLLQRMYNAELLEAEDILNYYNADTTDPVTLK----CKTFAEPFLQWL 181
+ + P +L+ +Y+ +L+E + IL + + V+ + AEPF++WL
Sbjct 316 KLHQTQLLPRVPIILKDLYDGDLVEEDVILAWAEKVSKKYVSKELAKEIHAKAEPFIKWL 375
Lambda K H
0.318 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4976880524
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40