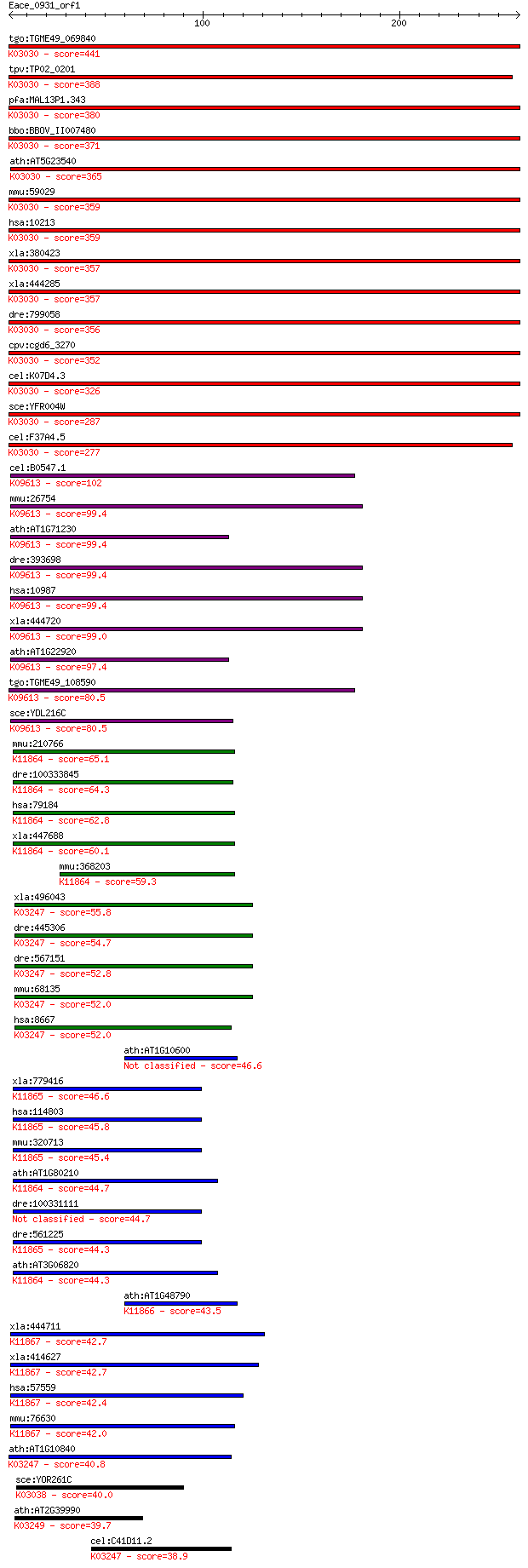
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0931_orf1
Length=260
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_069840 26S proteasome non-ATPase subunit, putative ... 441 1e-123
tpv:TP02_0201 proteasome regulatory subunit; K03030 26S protea... 388 1e-107
pfa:MAL13P1.343 proteasome regulatory subunit, putative; K0303... 380 2e-105
bbo:BBOV_II007480 18.m06620; 26S proteasome regulatory subunit... 371 2e-102
ath:AT5G23540 26S proteasome regulatory subunit, putative; K03... 365 1e-100
mmu:59029 Psmd14, 2610312C03Rik, 3200001M20Rik, AA986732, Pad1... 359 6e-99
hsa:10213 PSMD14, PAD1, POH1, RPN11; proteasome (prosome, macr... 359 6e-99
xla:380423 psmd14, MGC53911; 26S proteasome-associated pad1 ho... 357 2e-98
xla:444285 psmd14, MGC80929; proteasome (prosome, macropain) 2... 357 2e-98
dre:799058 psmd14, MGC162272, zgc:162272; proteasome (prosome,... 356 4e-98
cpv:cgd6_3270 26S proteasome-associated Mov34/MPN/PAD-1 family... 352 6e-97
cel:K07D4.3 rpn-11; proteasome Regulatory Particle, Non-ATPase... 326 5e-89
sce:YFR004W RPN11, MPR1; Metalloprotease subunit of the 19S re... 287 2e-77
cel:F37A4.5 hypothetical protein; K03030 26S proteasome regula... 277 3e-74
cel:B0547.1 csn-5; COP-9 SigNalosome subunit family member (cs... 102 1e-21
mmu:26754 Cops5, AI303502, CSN5, Jab1, Mov34, Sgn5; COP9 (cons... 99.4 1e-20
ath:AT1G71230 CSN5B; CSN5B (COP9-SIGNALOSOME 5B); protein bind... 99.4 1e-20
dre:393698 cops5, MGC73130, zgc:73130; COP9 constitutive photo... 99.4 1e-20
hsa:10987 COPS5, CSN5, JAB1, MGC3149, MOV-34, SGN5; COP9 const... 99.4 1e-20
xla:444720 cops5, MGC84682; COP9 constitutive photomorphogenic... 99.0 1e-20
ath:AT1G22920 CSN5A; CSN5A (COP9 SIGNALOSOME 5A); K09613 COP9 ... 97.4 4e-20
tgo:TGME49_108590 Mov34/MPN/PAD-1 domain-containing protein ; ... 80.5 5e-15
sce:YDL216C RRI1, CSN5, JAB1; Catalytic subunit of the COP9 si... 80.5 6e-15
mmu:210766 Brcc3, C6.1A, MGC29148; BRCA1/BRCA2-containing comp... 65.1 3e-10
dre:100333845 BRCA1/BRCA2-containing complex subunit 3-like; K... 64.3 4e-10
hsa:79184 BRCC3, BRCC36, C6.1A, CXorf53; BRCA1/BRCA2-containin... 62.8 1e-09
xla:447688 brcc3, MGC99130, brcc36; BRCA1/BRCA2-containing com... 60.1 7e-09
mmu:368203 Gm5136, C6.1AL, EG368203, MGC155743, MGC155745; pre... 59.3 2e-08
xla:496043 eif3h, eif3s3; eukaryotic translation initiation fa... 55.8 2e-07
dre:445306 eif3ha, eIF3h-A, eif3h, eif3s3, fj02c04, wu:fj02c04... 54.7 4e-07
dre:567151 eif3hb, MGC123049, zgc:123049; eukaryotic translati... 52.8 1e-06
mmu:68135 Eif3h, 1110008A16Rik, 40kD, 9430017H16Rik, EIF3-P40,... 52.0 2e-06
hsa:8667 EIF3H, EIF3S3, MGC102958, eIF3-gamma, eIF3-p40; eukar... 52.0 2e-06
ath:AT1G10600 hypothetical protein 46.6 9e-05
xla:779416 mysm1, 2A-DUB, MGC154322; myb-like, SWIRM and MPN d... 46.6 1e-04
hsa:114803 MYSM1, 2A-DUB, 2ADUB, DKFZp779J1554, DKFZp779J1721,... 45.8 1e-04
mmu:320713 Mysm1, C130067A03Rik, C530050H10Rik; myb-like, SWIR... 45.4 2e-04
ath:AT1G80210 hypothetical protein; K11864 BRCA1/BRCA2-contain... 44.7 3e-04
dre:100331111 histone H2A deubiquitinase MYSM1-like 44.7 4e-04
dre:561225 mysm1, im:7153144, si:ch211-59d15.8; Myb-like, SWIR... 44.3 4e-04
ath:AT3G06820 mov34 family protein; K11864 BRCA1/BRCA2-contain... 44.3 5e-04
ath:AT1G48790 mov34 family protein; K11866 STAM-binding protei... 43.5 7e-04
xla:444711 stambpl1, MGC84444; STAM binding protein-like 1 (EC... 42.7 0.001
xla:414627 hypothetical protein MGC81376; K11867 AMSH-like pro... 42.7 0.001
hsa:57559 STAMBPL1, ALMalpha, AMSH-FP, AMSH-LP, FLJ31524, KIAA... 42.4 0.002
mmu:76630 Stambpl1, 1700095N21Rik, 8230401J17Rik, ALMalpha, AM... 42.0 0.002
ath:AT1G10840 TIF3H1; TIF3H1; translation initiation factor; K... 40.8 0.005
sce:YOR261C RPN8; Essential, non-ATPase regulatory subunit of ... 40.0 0.009
ath:AT2G39990 EIF2; EIF2; translation initiation factor; K0324... 39.7 0.010
cel:C41D11.2 eif-3.H; Eukaryotic Initiation Factor family memb... 38.9 0.017
> tgo:TGME49_069840 26S proteasome non-ATPase subunit, putative
; K03030 26S proteasome regulatory subunit N11
Length=314
Score = 441 bits (1134), Expect = 1e-123, Method: Compositional matrix adjust.
Identities = 205/260 (78%), Positives = 233/260 (89%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEFIDD+TVRVVDVFSMPQSGNSVS+EAVD VYQTEML+QLKRTGRPEMVV
Sbjct 55 PMEVMGLMLGEFIDDYTVRVVDVFSMPQSGNSVSVEAVDPVYQTEMLEQLKRTGRPEMVV 114
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCWFSGTDVNTQQSFEQLN RAVGVVVDPIQSVKGKVV+DCFRLINPH LM
Sbjct 115 GWYHSHPGFGCWFSGTDVNTQQSFEQLNPRAVGVVVDPIQSVKGKVVIDCFRLINPHLLM 174
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LGQE RQTTSNIGHLQRPTI+ALVHGLNRNYY+I IN RK ELE+QML NLH+NKWNDAL
Sbjct 175 LGQELRQTTSNIGHLQRPTISALVHGLNRNYYAIVINYRKNELENQMLLNLHRNKWNDAL 234
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
KL+ F+E E A ++K+LS +Y M++EEIKK PEQL+V+RAG++DAK RL+N +D
Sbjct 235 KLKPFDEMAAESAACTKSMKELSEQYNKMVQEEIKKTPEQLVVERAGKVDAKKRLENDVD 294
Query 241 SLLTDTLLQNLGSMISTLAF 260
+L+T+ +L +LG+MI TL F
Sbjct 295 TLMTENILHSLGTMIDTLVF 314
> tpv:TP02_0201 proteasome regulatory subunit; K03030 26S proteasome
regulatory subunit N11
Length=312
Score = 388 bits (997), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 178/256 (69%), Positives = 213/256 (83%), Gaps = 0/256 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLG+FIDD+T+RVVDVFSMPQSGNSVS+EAVD VYQTEM DQLKRTGRPE+VV
Sbjct 53 PMEVMGLMLGDFIDDYTIRVVDVFSMPQSGNSVSVEAVDPVYQTEMKDQLKRTGRPEVVV 112
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCWFSGTDVNTQQSFEQLN RAVGVV+DPIQSVKGKVV+DCFRLI+PH +M
Sbjct 113 GWYHSHPGFGCWFSGTDVNTQQSFEQLNPRAVGVVIDPIQSVKGKVVIDCFRLISPHLIM 172
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LG EPRQTTSNIGHLQ+PTI ALVHGLNRNYYSI INC+K LE QML N +KN+W L
Sbjct 173 LGHEPRQTTSNIGHLQKPTIIALVHGLNRNYYSIVINCKKTPLESQMLLNFNKNRWTKDL 232
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
LQ+F E +E V ++DL KY I++E+ KPE+L+V G++DAK ++N+++
Sbjct 233 HLQDFVERQKENNDLVREIRDLCEKYNQSIKQEMTCKPEELVVANVGKLDAKKHIENSVN 292
Query 241 SLLTDTLLQNLGSMIS 256
+LL++ L +M++
Sbjct 293 TLLSNNTLNVFSTMLA 308
> pfa:MAL13P1.343 proteasome regulatory subunit, putative; K03030
26S proteasome regulatory subunit N11
Length=311
Score = 380 bits (977), Expect = 2e-105, Method: Compositional matrix adjust.
Identities = 172/260 (66%), Positives = 215/260 (82%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGE +D++T+R+VDVF+MPQSGNSVS+EAVD VYQT ML++LK+TGR EMVV
Sbjct 52 PMEVMGLMLGEIVDEYTIRIVDVFAMPQSGNSVSVEAVDPVYQTNMLEELKKTGRHEMVV 111
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SGTDVNTQ+SFEQLN R +GVVVDPIQSVKGKVV+DCFRLINPH LM
Sbjct 112 GWYHSHPGFGCWLSGTDVNTQKSFEQLNPRTIGVVVDPIQSVKGKVVIDCFRLINPHILM 171
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LGQEPRQTTSNIG+L +PT+TALVHGLNRNYYSI IN RK ELE ML NLHK+ W + L
Sbjct 172 LGQEPRQTTSNIGYLTKPTLTALVHGLNRNYYSIVINYRKNELEKNMLLNLHKDMWTNPL 231
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
KL +F E + + ++ +K L+ Y + E+KK E++L++ G+IDAK R+QN+++
Sbjct 232 KLNDFHEQKKSSDETLEDIKKLTTLYNKNLRNEMKKTSEEILLENIGKIDAKKRIQNSVE 291
Query 241 SLLTDTLLQNLGSMISTLAF 260
+LL +++L +G+M +TL F
Sbjct 292 TLLNESILTCIGTMANTLFF 311
> bbo:BBOV_II007480 18.m06620; 26S proteasome regulatory subunit;
K03030 26S proteasome regulatory subunit N11
Length=312
Score = 371 bits (952), Expect = 2e-102, Method: Compositional matrix adjust.
Identities = 171/260 (65%), Positives = 205/260 (78%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEFIDD+T+ VVDVFSMPQSGNSVS+EAVD VYQTEM D+LK TGRPE+VV
Sbjct 53 PMEVMGLMLGEFIDDYTIVVVDVFSMPQSGNSVSVEAVDPVYQTEMKDKLKLTGRPEVVV 112
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCWFSGTD+NTQQSFEQLN RAVG+V+DPIQSVKGKVV+DCFRLI PH +M
Sbjct 113 GWYHSHPGFGCWFSGTDINTQQSFEQLNPRAVGIVIDPIQSVKGKVVIDCFRLITPHLIM 172
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LGQEPRQTTSNIGHL +PT+ A+VHGLNRNYY+I IN RK LE QML N H+NKW D L
Sbjct 173 LGQEPRQTTSNIGHLSKPTMIAVVHGLNRNYYNIVINYRKSVLETQMLMNYHRNKWTDNL 232
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
++++F RE +V +KDL KY I++E+ ++L V G+++AK + N +
Sbjct 233 QVRDFVTRRRENRETVGNIKDLIDKYNDSIKQELTSTADELAVANVGKLNAKAHIDNHVS 292
Query 241 SLLTDTLLQNLGSMISTLAF 260
LL D L G+M++ F
Sbjct 293 RLLKDNSLDTFGTMLAVEMF 312
> ath:AT5G23540 26S proteasome regulatory subunit, putative; K03030
26S proteasome regulatory subunit N11
Length=259
Score = 365 bits (936), Expect = 1e-100, Method: Compositional matrix adjust.
Identities = 166/259 (64%), Positives = 205/259 (79%), Gaps = 0/259 (0%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVG 61
MEVMGLMLGEF+D++TVRVVDVF+MPQSG VS+EAVD+V+QT MLD LK+TGRPEMVVG
Sbjct 1 MEVMGLMLGEFVDEYTVRVVDVFAMPQSGTGVSVEAVDHVFQTNMLDMLKQTGRPEMVVG 60
Query 62 WYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLML 121
WYHSHPGFGCW SG D+NTQQSFE LNQRAV VVVDPIQSVKGKVV+D FR INP +ML
Sbjct 61 WYHSHPGFGCWLSGVDINTQQSFEALNQRAVAVVVDPIQSVKGKVVIDAFRSINPQTIML 120
Query 122 GQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDALK 181
GQEPRQTTSN+GHL +P+I AL+HGLNR+YYSI IN RK ELE +ML NLHK KW D L
Sbjct 121 GQEPRQTTSNLGHLNKPSIQALIHGLNRHYYSIAINYRKNELEEKMLLNLHKKKWTDGLT 180
Query 182 LQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTIDS 241
L+ F+ H + E +V + L+AKY ++EE + PE+L + G+ DAK L+ + +
Sbjct 181 LRRFDTHSKTNEQTVQEMLSLAAKYNKAVQEEDELSPEKLAIVNVGRQDAKKHLEEHVSN 240
Query 242 LLTDTLLQNLGSMISTLAF 260
L++ ++Q LG+M+ T+ F
Sbjct 241 LMSSNIVQTLGTMLDTVVF 259
> mmu:59029 Psmd14, 2610312C03Rik, 3200001M20Rik, AA986732, Pad1,
Poh1, rpm11; proteasome (prosome, macropain) 26S subunit,
non-ATPase, 14 (EC:3.1.2.15); K03030 26S proteasome regulatory
subunit N11
Length=310
Score = 359 bits (921), Expect = 6e-99, Method: Compositional matrix adjust.
Identities = 163/260 (62%), Positives = 203/260 (78%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TVRV+DVF+MPQSG VS+EAVD V+Q +MLD LK+TGRPEMVV
Sbjct 50 PMEVMGLMLGEFVDDYTVRVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQTGRPEMVV 109
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SG D+NTQQSFE L++RAV VVVDPIQSVKGKVV+D FRLIN + ++
Sbjct 110 GWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVVIDAFRLINANMMV 169
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LG EPRQTTSN+GHL +P+I AL+HGLNR+YYSITIN RK ELE +ML NLHK W + L
Sbjct 170 LGHEPRQTTSNLGHLNKPSIQALIHGLNRHYYSITINYRKNELEQKMLLNLHKKSWMEGL 229
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
LQ++ EH + E+ V + +L+ Y +EEE K PEQL + G+ D K L+ +D
Sbjct 230 TLQDYSEHCKHNESVVKEMLELAKNYNKAVEEEDKMTPEQLAIKNVGKQDPKRHLEEHVD 289
Query 241 SLLTDTLLQNLGSMISTLAF 260
L+T ++Q L +M+ T+ F
Sbjct 290 VLMTSNIVQCLAAMLDTVVF 309
> hsa:10213 PSMD14, PAD1, POH1, RPN11; proteasome (prosome, macropain)
26S subunit, non-ATPase, 14 (EC:3.1.2.15); K03030 26S
proteasome regulatory subunit N11
Length=310
Score = 359 bits (921), Expect = 6e-99, Method: Compositional matrix adjust.
Identities = 163/260 (62%), Positives = 203/260 (78%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TVRV+DVF+MPQSG VS+EAVD V+Q +MLD LK+TGRPEMVV
Sbjct 50 PMEVMGLMLGEFVDDYTVRVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQTGRPEMVV 109
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SG D+NTQQSFE L++RAV VVVDPIQSVKGKVV+D FRLIN + ++
Sbjct 110 GWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVVIDAFRLINANMMV 169
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LG EPRQTTSN+GHL +P+I AL+HGLNR+YYSITIN RK ELE +ML NLHK W + L
Sbjct 170 LGHEPRQTTSNLGHLNKPSIQALIHGLNRHYYSITINYRKNELEQKMLLNLHKKSWMEGL 229
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
LQ++ EH + E+ V + +L+ Y +EEE K PEQL + G+ D K L+ +D
Sbjct 230 TLQDYSEHCKHNESVVKEMLELAKNYNKAVEEEDKMTPEQLAIKNVGKQDPKRHLEEHVD 289
Query 241 SLLTDTLLQNLGSMISTLAF 260
L+T ++Q L +M+ T+ F
Sbjct 290 VLMTSNIVQCLAAMLDTVVF 309
> xla:380423 psmd14, MGC53911; 26S proteasome-associated pad1
homolog (EC:3.1.2.15); K03030 26S proteasome regulatory subunit
N11
Length=310
Score = 357 bits (916), Expect = 2e-98, Method: Compositional matrix adjust.
Identities = 163/260 (62%), Positives = 202/260 (77%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TVRV+DVF+MPQSG VS+EAVD V+Q +MLD LK+TGRPEMVV
Sbjct 50 PMEVMGLMLGEFVDDYTVRVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQTGRPEMVV 109
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SG D+NTQQSFE L++RAV VVVDPIQSVKGKVV+D FRLIN + ++
Sbjct 110 GWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVVIDAFRLINANMMV 169
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LG EPRQTTSN+GHL +P+I AL+HGLNR+YYSITIN RK ELE +ML NLHK W + L
Sbjct 170 LGHEPRQTTSNLGHLNKPSIQALIHGLNRHYYSITINYRKNELEQKMLLNLHKKSWMEGL 229
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
LQ++ EH + E V + +L+ Y +EEE K PEQL + G+ D K L+ +D
Sbjct 230 TLQDYSEHCKLNETVVKEMLELAKNYNKAVEEEDKMTPEQLAIKNVGKQDPKRHLEEHVD 289
Query 241 SLLTDTLLQNLGSMISTLAF 260
L+T ++Q L +M+ T+ F
Sbjct 290 VLMTSNIVQCLAAMLDTVVF 309
> xla:444285 psmd14, MGC80929; proteasome (prosome, macropain)
26S subunit, non-ATPase, 14; K03030 26S proteasome regulatory
subunit N11
Length=310
Score = 357 bits (916), Expect = 2e-98, Method: Compositional matrix adjust.
Identities = 163/260 (62%), Positives = 202/260 (77%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TVRV+DVF+MPQSG VS+EAVD V+Q +MLD LK+TGRPEMVV
Sbjct 50 PMEVMGLMLGEFVDDYTVRVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQTGRPEMVV 109
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SG D+NTQQSFE L++RAV VVVDPIQSVKGKVV+D FRLIN + ++
Sbjct 110 GWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVVIDAFRLINANMMV 169
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LG EPRQTTSN+GHL +P+I AL+HGLNR+YYSITIN RK ELE +ML NLHK W + L
Sbjct 170 LGHEPRQTTSNLGHLNKPSIQALIHGLNRHYYSITINYRKNELEQKMLLNLHKKSWMEGL 229
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
LQ++ EH + E V + +L+ Y +EEE K PEQL + G+ D K L+ +D
Sbjct 230 TLQDYSEHCKLNETVVKEMLELAKNYNKAVEEEDKMTPEQLAIKNVGKQDPKRHLEEHVD 289
Query 241 SLLTDTLLQNLGSMISTLAF 260
L+T ++Q L +M+ T+ F
Sbjct 290 VLMTSNIVQCLAAMLDTVVF 309
> dre:799058 psmd14, MGC162272, zgc:162272; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 14 (EC:3.1.2.15); K03030
26S proteasome regulatory subunit N11
Length=310
Score = 356 bits (914), Expect = 4e-98, Method: Compositional matrix adjust.
Identities = 163/260 (62%), Positives = 202/260 (77%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TVRV+DVF+MPQSG VS+EAVD V+Q +MLD LK+TGRPEMVV
Sbjct 50 PMEVMGLMLGEFVDDYTVRVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQTGRPEMVV 109
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SG D+NTQQSFE L++RAV VVVDPIQSVKGKVV+D FRLIN + ++
Sbjct 110 GWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVVIDAFRLINANMMV 169
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
LG EPRQTTSN+GHL +P+I AL+HGLNR+YYSITIN RK ELE +ML NLHK W + L
Sbjct 170 LGHEPRQTTSNLGHLNKPSIQALIHGLNRHYYSITINYRKNELEQKMLLNLHKKSWMEGL 229
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
LQ++ EH + E V + +L+ Y +EEE K PEQL + G+ D K L+ +D
Sbjct 230 TLQDYSEHCKLNETIVKEMLELAKNYNKAVEEEDKMTPEQLAIKNVGKQDPKRHLEEHVD 289
Query 241 SLLTDTLLQNLGSMISTLAF 260
L+T ++Q L +M+ T+ F
Sbjct 290 VLMTSNIVQCLAAMLDTVVF 309
> cpv:cgd6_3270 26S proteasome-associated Mov34/MPN/PAD-1 family.
JAB domain. ; K03030 26S proteasome regulatory subunit N11
Length=315
Score = 352 bits (904), Expect = 6e-97, Method: Compositional matrix adjust.
Identities = 166/260 (63%), Positives = 204/260 (78%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGL+LGEFIDD++VRVVDVFSMPQSGNSVS+EAVD VYQT+ML+ LKR GR E+VV
Sbjct 56 PMEVMGLLLGEFIDDYSVRVVDVFSMPQSGNSVSVEAVDPVYQTDMLEMLKRVGRSELVV 115
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCWFSGTDV+TQQSFEQLN RAVG+VVDPIQSVKGKVV+DCFRLI+PH ++
Sbjct 116 GWYHSHPGFGCWFSGTDVSTQQSFEQLNPRAVGIVVDPIQSVKGKVVIDCFRLISPHSVI 175
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
GQEPRQTTSNIGHLQ+P+ITALVHGLNRNYYSI I RK LE +ML NLHK W++ L
Sbjct 176 AGQEPRQTTSNIGHLQKPSITALVHGLNRNYYSIAIRYRKNLLEQKMLLNLHKPTWSEPL 235
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
+ E + + + + + S +Y + + K E+ ++ G+IDAK RL ++
Sbjct 236 RCDKEENFNERNNSMIKRICETSKQYHESTKLGLSKTSEEFELENVGKIDAKKRLNADVE 295
Query 241 SLLTDTLLQNLGSMISTLAF 260
++LTD +LQ L S I++ F
Sbjct 296 TVLTDNILQILKSNIASSVF 315
> cel:K07D4.3 rpn-11; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-11); K03030 26S proteasome regulatory
subunit N11
Length=312
Score = 326 bits (836), Expect = 5e-89, Method: Compositional matrix adjust.
Identities = 147/260 (56%), Positives = 194/260 (74%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TV V+DVF+MPQSG VS+EAVD V+Q +MLD LK+TGRPEMVV
Sbjct 52 PMEVMGLMLGEFVDDYTVNVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQTGRPEMVV 111
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW SG D+NTQQSFE L+ RAV VVVDPIQSVKGKVV+D FR INP +
Sbjct 112 GWYHSHPGFGCWLSGVDINTQQSFEALSDRAVAVVVDPIQSVKGKVVIDAFRTINPQSMA 171
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
L QEPRQTTSN+GHLQ+P+I AL+HGLNR+YYSI I R +LE +ML NL+K W DA+
Sbjct 172 LNQEPRQTTSNLGHLQKPSIQALIHGLNRHYYSIPIAYRTHDLEQKMLLNLNKLSWMDAV 231
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
++N+ + + + + A+ L+ Y +E+E ++L + G++D K + + +
Sbjct 232 SVENYSKCGEQNKEHLKAMLKLAKNYKKALEDEKNMTDQELAIKNVGKMDPKRHIADEVS 291
Query 241 SLLTDTLLQNLGSMISTLAF 260
+L D ++Q+L M++T +
Sbjct 292 KMLNDNIVQSLAGMMATTSL 311
> sce:YFR004W RPN11, MPR1; Metalloprotease subunit of the 19S
regulatory particle of the 26S proteasome lid; couples the deubiquitination
and degradation of proteasome substrates; involved,
independent of catalytic activity, in fission of mitochondria
and peroxisomes; K03030 26S proteasome regulatory
subunit N11
Length=306
Score = 287 bits (735), Expect = 2e-77, Method: Compositional matrix adjust.
Identities = 143/260 (55%), Positives = 183/260 (70%), Gaps = 0/260 (0%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
PMEVMGLMLGEF+DD+TV VVDVF+MPQSG VS+EAVD V+Q +M+D LK+TGR +MVV
Sbjct 46 PMEVMGLMLGEFVDDYTVNVVDVFAMPQSGTGVSVEAVDDVFQAKMMDMLKQTGRDQMVV 105
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW S DVNTQ+SFEQLN RAV VVVDPIQSVKGKVV+D FRLI+ L+
Sbjct 106 GWYHSHPGFGCWLSSVDVNTQKSFEQLNSRAVAVVVDPIQSVKGKVVIDAFRLIDTGALI 165
Query 121 LGQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNKWNDAL 180
EPRQTTSN G L + I AL+HGLNR+YYS+ I+ K E +ML NLHK +W L
Sbjct 166 NNLEPRQTTSNTGLLNKANIQALIHGLNRHYYSLNIDYHKTAKETKMLMNLHKEQWQSGL 225
Query 181 KLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKPEQLLVDRAGQIDAKMRLQNTID 240
K+ ++EE A+ ++ ++ +Y IEEE + E+L G+ D K L T D
Sbjct 226 KMYDYEEKEESNLAATKSMVKIAEQYSKRIEEEKELTEEELKTRYVGRQDPKKHLSETAD 285
Query 241 SLLTDTLLQNLGSMISTLAF 260
L + ++ L + ++++A
Sbjct 286 ETLENNIVSVLTAGVNSVAI 305
> cel:F37A4.5 hypothetical protein; K03030 26S proteasome regulatory
subunit N11
Length=319
Score = 277 bits (708), Expect = 3e-74, Method: Compositional matrix adjust.
Identities = 138/268 (51%), Positives = 182/268 (67%), Gaps = 13/268 (4%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
P+EVMGLMLG+F+DD+T+ V DVF+MPQSG SV++E+VD VYQT+ +D LK GR E VV
Sbjct 48 PLEVMGLMLGDFVDDYTINVTDVFAMPQSGTSVTVESVDPVYQTKHMDLLKLVGRTENVV 107
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GWYHSHPGFGCW S DVNTQQSFE L+ RAV VVVDPIQSVKGKV++D FR +NP L
Sbjct 108 GWYHSHPGFGCWLSSVDVNTQQSFEALHPRAVAVVVDPIQSVKGKVMLDAFRSVNPLNLQ 167
Query 121 L-----GQEPRQTTSNIGHLQRPTITALVHGLNRNYYSITINCRKGELEHQMLANLHKNK 175
+ EPRQTTSN+GHL +P++ ++VHGL YYS+ + R G E +ML L+K
Sbjct 168 IRPLAPTAEPRQTTSNLGHLTKPSLISVVHGLGTKYYSLNVAYRMGSNEQKMLMCLNKKS 227
Query 176 WNDALKLQNFEEHHREREASVAALKDLSAKYISMIEEEIKKKP-------EQLLVDRAGQ 228
W D L + + E +++E ++ L A + I +E+K+KP Q V + G+
Sbjct 228 WYDQLNMSTYSELEKKQEEKFKSINKLIAVFNKDI-DEVKEKPIADKKGKTQEEVKKFGK 286
Query 229 IDAKMRLQNTIDSLLTDTLLQNLGSMIS 256
I+AK +LQ SLL D+L L +MI+
Sbjct 287 INAKQQLQMITSSLLNDSLCHQLTAMIN 314
> cel:B0547.1 csn-5; COP-9 SigNalosome subunit family member (csn-5);
K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=368
Score = 102 bits (255), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 61/179 (34%), Positives = 96/179 (53%), Gaps = 11/179 (6%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNS--VSIEAVDYVYQTEMLDQLKRTGRPEMV 59
+E+MGL+ G ID + ++DVF++P G V+ +A Y Y T + GR E V
Sbjct 76 LEIMGLLQGR-IDANSFIILDVFALPVEGTETRVNAQAQAYEYMTVYSEMCDTEGRKEKV 134
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFRLINPHF 118
VGWYHSHPG+GCW SG DV+TQ ++ + V +V+DP++++ GKV + FR +
Sbjct 135 VGWYHSHPGYGCWLSGIDVSTQTLNQKFQEPWVAIVIDPLRTMSAGKVDIGAFRTYPEGY 194
Query 119 LMLGQEPRQTTSNIGHLQRPTITAL-VHGLNRNYYSITINCRKGELEHQMLANLHKNKW 176
+ P S + I VH + YYS+ ++ K +L+ +L +L + W
Sbjct 195 ----RPPDDVPSEYQSIPLAKIEDFGVHC--KRYYSLDVSFFKSQLDAHILTSLWNSYW 247
> mmu:26754 Cops5, AI303502, CSN5, Jab1, Mov34, Sgn5; COP9 (constitutive
photomorphogenic) homolog, subunit 5 (Arabidopsis
thaliana); K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=334
Score = 99.4 bits (246), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 63/183 (34%), Positives = 99/183 (54%), Gaps = 11/183 (6%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNS--VSIEAVDYVYQTEMLDQLKRTGRPEMV 59
+EVMGLMLG+ +D T+ ++D F++P G V+ +A Y Y ++ K+ GR E
Sbjct 75 LEVMGLMLGK-VDGETMIIMDSFALPVEGTETRVNAQAAAYEYMAAYIENAKQVGRLENA 133
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFRLINPHF 118
+GWYHSHPG+GCW SG DV+TQ +Q + V VV+DP +++ GKV + FR +
Sbjct 134 IGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKVNLGAFRTYPKGY 193
Query 119 LMLGQEPRQTTSNIGHLQRPTITAL-VHGLNRNYYSITINCRKGELEHQMLANLHKNKWN 177
+ P + S + I VH + YY++ ++ K L+ ++L L W
Sbjct 194 ----KPPDEGPSEYQTIPLNKIEDFGVHC--KQYYALEVSYFKSSLDRKLLELLWNKYWV 247
Query 178 DAL 180
+ L
Sbjct 248 NTL 250
> ath:AT1G71230 CSN5B; CSN5B (COP9-SIGNALOSOME 5B); protein binding;
K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=358
Score = 99.4 bits (246), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 52/114 (45%), Positives = 70/114 (61%), Gaps = 4/114 (3%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVD--YVYQTEMLDQLKRTGRPEMV 59
+E+MGLM G+ D T+ V+D F++P G + A D Y Y E K GR E V
Sbjct 79 IEIMGLMQGK-TDGDTIIVMDAFALPVEGTETRVNAQDDAYEYMVEYSQTNKLAGRLENV 137
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFR 112
VGWYHSHPG+GCW SG DV+TQ+ +Q + + VV+DP ++V GKV + FR
Sbjct 138 VGWYHSHPGYGCWLSGIDVSTQRLNQQHQEPFLAVVIDPTRTVSAGKVEIGAFR 191
> dre:393698 cops5, MGC73130, zgc:73130; COP9 constitutive photomorphogenic
homolog subunit 5; K09613 COP9 signalosome complex
subunit 5 [EC:3.4.-.-]
Length=334
Score = 99.4 bits (246), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 63/183 (34%), Positives = 99/183 (54%), Gaps = 11/183 (6%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNS--VSIEAVDYVYQTEMLDQLKRTGRPEMV 59
+EVMGLMLG+ +D T+ ++D F++P G V+ +A Y Y ++ K+ GR E
Sbjct 73 LEVMGLMLGK-VDGETMIIMDSFALPVEGTETRVNAQAAAYEYMAAYIENAKQVGRLENA 131
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFRLINPHF 118
+GWYHSHPG+GCW SG DV+TQ +Q + V VV+DP +++ GKV + FR +
Sbjct 132 IGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKVNLGAFRTYPKGY 191
Query 119 LMLGQEPRQTTSNIGHLQRPTITAL-VHGLNRNYYSITINCRKGELEHQMLANLHKNKWN 177
+ P + S + I VH + YY++ ++ K L+ ++L L W
Sbjct 192 ----KPPDEGPSEYQTIPLNKIEDFGVHC--KQYYALEVSYFKSSLDRKLLELLWNKYWV 245
Query 178 DAL 180
+ L
Sbjct 246 NTL 248
> hsa:10987 COPS5, CSN5, JAB1, MGC3149, MOV-34, SGN5; COP9 constitutive
photomorphogenic homolog subunit 5 (Arabidopsis);
K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=334
Score = 99.4 bits (246), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 63/183 (34%), Positives = 99/183 (54%), Gaps = 11/183 (6%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNS--VSIEAVDYVYQTEMLDQLKRTGRPEMV 59
+EVMGLMLG+ +D T+ ++D F++P G V+ +A Y Y ++ K+ GR E
Sbjct 75 LEVMGLMLGK-VDGETMIIMDSFALPVEGTETRVNAQAAAYEYMAAYIENAKQVGRLENA 133
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFRLINPHF 118
+GWYHSHPG+GCW SG DV+TQ +Q + V VV+DP +++ GKV + FR +
Sbjct 134 IGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKVNLGAFRTYPKGY 193
Query 119 LMLGQEPRQTTSNIGHLQRPTITAL-VHGLNRNYYSITINCRKGELEHQMLANLHKNKWN 177
+ P + S + I VH + YY++ ++ K L+ ++L L W
Sbjct 194 ----KPPDEGPSEYQTIPLNKIEDFGVHC--KQYYALEVSYFKSSLDRKLLELLWNKYWV 247
Query 178 DAL 180
+ L
Sbjct 248 NTL 250
> xla:444720 cops5, MGC84682; COP9 constitutive photomorphogenic
homolog subunit 5; K09613 COP9 signalosome complex subunit
5 [EC:3.4.-.-]
Length=332
Score = 99.0 bits (245), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 63/183 (34%), Positives = 98/183 (53%), Gaps = 11/183 (6%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNS--VSIEAVDYVYQTEMLDQLKRTGRPEMV 59
+EVMGLMLG+ +D T+ ++D F++P G V+ +A Y Y ++ K+ GR E
Sbjct 73 LEVMGLMLGK-VDGETMIIMDSFALPVEGTETRVNAQAAAYEYMAAYIENAKQVGRLENA 131
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFRLINPHF 118
+GWYHSHPG+GCW SG DV+TQ +Q + V VV+DP +++ GKV + FR +
Sbjct 132 IGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKVNLGAFRTYPKGY 191
Query 119 LMLGQEPRQTTSNIGHLQRPTITAL-VHGLNRNYYSITINCRKGELEHQMLANLHKNKWN 177
+ P + S + I VH + YY++ + K L+ ++L L W
Sbjct 192 ----KPPDEGPSEYQTIPLNKIEDFGVHC--KQYYALEVTYFKSSLDRKLLELLWNKYWV 245
Query 178 DAL 180
+ L
Sbjct 246 NTL 248
> ath:AT1G22920 CSN5A; CSN5A (COP9 SIGNALOSOME 5A); K09613 COP9
signalosome complex subunit 5 [EC:3.4.-.-]
Length=357
Score = 97.4 bits (241), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 51/114 (44%), Positives = 68/114 (59%), Gaps = 4/114 (3%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVD--YVYQTEMLDQLKRTGRPEMV 59
+E+MGLM G+ D T+ V+D F++P G + A Y Y E K GR E V
Sbjct 79 IEIMGLMQGKTEGD-TIIVMDAFALPVEGTETRVNAQSDAYEYMVEYSQTSKLAGRLENV 137
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVK-GKVVMDCFR 112
VGWYHSHPG+GCW SG DV+TQ +Q + + VV+DP ++V GKV + FR
Sbjct 138 VGWYHSHPGYGCWLSGIDVSTQMLNQQYQEPFLAVVIDPTRTVSAGKVEIGAFR 191
> tgo:TGME49_108590 Mov34/MPN/PAD-1 domain-containing protein
; K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=321
Score = 80.5 bits (197), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 65/212 (30%), Positives = 92/212 (43%), Gaps = 41/212 (19%)
Query 1 PMEVMGLMLG----------------EFIDDFTVRVVDVFSMPQSGNSVSIEA------- 37
P+EVMGLMLG E + D V VF +P G + A
Sbjct 71 PLEVMGLMLGSVSPISSSVKSASSPLEQLGDQAFVVHSVFRLPVEGTETRVNAGAEANEY 130
Query 38 -VDYVYQTE-MLDQLKRTGRPE-----MVVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQR 90
VD+V + E L R P+ VVGWYHSHPG+ CW SG DV TQ+ ++
Sbjct 131 MVDFVQRAEEALSPPVRPDEPDEGVGLCVVGWYHSHPGYRCWLSGIDVETQKLHQRGQDP 190
Query 91 AVGVVVDPIQSVK-GKVVMDCFRLINPHF----LMLGQEPRQTTSNIGHLQRPTITALVH 145
V VVVDP +++ G+V + FR H L Q+ R + + P A
Sbjct 191 FVAVVVDPTRTLATGEVDIGAFRCYPDHSQNARLDSVQKSRHDQAGV-----PAEKASDF 245
Query 146 GLN-RNYYSITINCRKGELEHQMLANLHKNKW 176
G++ R YY + + L+ ++ L + W
Sbjct 246 GVHWREYYKLNVELLCSSLDALLIERLSEAAW 277
> sce:YDL216C RRI1, CSN5, JAB1; Catalytic subunit of the COP9
signalosome (CSN) complex that acts as an isopeptidase in cleaving
the ubiquitin-like protein Nedd8 from SCF ubiquitin ligases;
metalloendopeptidase involved in the adaptation to pheromone
signaling (EC:3.4.-.-); K09613 COP9 signalosome complex
subunit 5 [EC:3.4.-.-]
Length=440
Score = 80.5 bits (197), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 68/126 (53%), Gaps = 14/126 (11%)
Query 2 MEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEA--VDYVYQTEMLDQL--------- 50
+E+MG+++G + D V V+D F++P G + A Y Y + +D++
Sbjct 91 IEIMGILMGFTLKDNIV-VMDCFNLPVVGTETRVNAQLESYEYMVQYIDEMYNHNDGGDG 149
Query 51 -KRTGRPEMVVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVV-M 108
G VVGW+HSHPG+ CW S D+ TQ ++ V +VVDP++S++ K++ M
Sbjct 150 RDYKGAKLNVVGWFHSHPGYDCWLSNIDIQTQDLNQRFQDPYVAIVVDPLKSLEDKILRM 209
Query 109 DCFRLI 114
FR I
Sbjct 210 GAFRTI 215
> mmu:210766 Brcc3, C6.1A, MGC29148; BRCA1/BRCA2-containing complex,
subunit 3 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=291
Score = 65.1 bits (157), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/154 (30%), Positives = 68/154 (44%), Gaps = 50/154 (32%)
Query 3 EVMGLMLGEFIDDF---------------------TVRVVDVFSM--------------- 26
EVMGL +GE DD T+R+V + S+
Sbjct 33 EVMGLCIGELNDDIRSDSKFTYTGTEMRTVQEKMDTIRIVHIHSVIILRRSDKRKDRVEI 92
Query 27 -PQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGWYHSHPGFGCWFSGTDVNTQQSFE 85
P+ ++ S EA E L +L TGRP VVGWYHSHP W S DV TQ ++
Sbjct 93 SPEQLSAASTEA-------ERLAEL--TGRPMRVVGWYHSHPHITVWPSHVDVRTQAMYQ 143
Query 86 QLNQRAVGVV----VDPIQSVKGKVVMDCFRLIN 115
++Q VG++ ++ + G+V+ CF+ I
Sbjct 144 MMDQGFVGLIFSCFIEDKNTKTGRVLYTCFQSIQ 177
> dre:100333845 BRCA1/BRCA2-containing complex subunit 3-like;
K11864 BRCA1/BRCA2-containing complex subunit 3
Length=260
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 61/121 (50%), Gaps = 10/121 (8%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSMPQSGN-----SVSIEAVDYVYQTEMLDQLKRTGRPE 57
EVMGL +GE + V + V + +S +S E + TE + TGRP
Sbjct 27 EVMGLCIGEVDTNRIVHIHSVIILRRSDKRKDRVEISPEQLSAA-STEAERLAEMTGRPM 85
Query 58 MVVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVV----VDPIQSVKGKVVMDCFRL 113
VVGWYHSHP W S DV TQ ++ ++Q VG++ ++ + G+V+ CF+
Sbjct 86 RVVGWYHSHPHITVWPSHVDVRTQAMYQMMDQGFVGLIFSCFIEDKNTKTGRVLYTCFQS 145
Query 114 I 114
+
Sbjct 146 V 146
> hsa:79184 BRCC3, BRCC36, C6.1A, CXorf53; BRCA1/BRCA2-containing
complex, subunit 3 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=291
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 47/154 (30%), Positives = 67/154 (43%), Gaps = 50/154 (32%)
Query 3 EVMGLMLGEFIDDF---------------------TVRVVDVFSM--------------- 26
EVMGL +GE DD VR+V + S+
Sbjct 33 EVMGLCIGELNDDTRSDSKFAYTGTEMRTVAEKVDAVRIVHIHSVIILRRSDKRKDRVEI 92
Query 27 -PQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGWYHSHPGFGCWFSGTDVNTQQSFE 85
P+ ++ S EA E L +L TGRP VVGWYHSHP W S DV TQ ++
Sbjct 93 SPEQLSAASTEA-------ERLAEL--TGRPMRVVGWYHSHPHITVWPSHVDVRTQAMYQ 143
Query 86 QLNQRAVGVV----VDPIQSVKGKVVMDCFRLIN 115
++Q VG++ ++ + G+V+ CF+ I
Sbjct 144 MMDQGFVGLIFSCFIEDKNTKTGRVLYTCFQSIQ 177
> xla:447688 brcc3, MGC99130, brcc36; BRCA1/BRCA2-containing complex,
subunit 3 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=261
Score = 60.1 bits (144), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 59/121 (48%), Gaps = 8/121 (6%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSMPQSG---NSVSIEAVDYVYQTEMLDQLKR-TGRPEM 58
EVMGL +GE V + V + +S + V I T D+L TGRP
Sbjct 27 EVMGLCIGEVDTQKLVHIHSVIILRRSDKRKDRVEISPEQLSAATIEADRLADITGRPMR 86
Query 59 VVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVV----VDPIQSVKGKVVMDCFRLI 114
VVGWYHSHP W S DV TQ ++ ++ VG++ ++ + G+++ CF+ +
Sbjct 87 VVGWYHSHPHITVWPSHVDVRTQAMYQMMDVGFVGLIFSCFIEDKNTKTGRILYTCFQSV 146
Query 115 N 115
Sbjct 147 Q 147
> mmu:368203 Gm5136, C6.1AL, EG368203, MGC155743, MGC155745; predicted
gene 5136 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=291
Score = 59.3 bits (142), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 13/93 (13%)
Query 27 PQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGWYHSHPGFGCWFSGTDVNTQQSFEQ 86
P+ ++ SIEA QT GRP VVGWYHSHP W S DV TQ ++
Sbjct 94 PEQLSAASIEAERLAEQT---------GRPMRVVGWYHSHPHITVWPSHVDVRTQAMYQM 144
Query 87 LNQRAVGVV----VDPIQSVKGKVVMDCFRLIN 115
++Q VG++ ++ + G+V+ CF+ +
Sbjct 145 MDQSFVGLIFACFIEDKPTKIGRVLYTCFQSVQ 177
> xla:496043 eif3h, eif3s3; eukaryotic translation initiation
factor 3, subunit H; K03247 translation initiation factor 3
subunit H
Length=334
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 66/122 (54%), Gaps = 6/122 (4%)
Query 4 VMGLMLGEFIDDFTVRVVDVFSMPQ-SGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGW 62
V G++LG +DD + + + F PQ + + V + V Y Q EM+ L+ + VGW
Sbjct 45 VQGVLLGLVVDD-RLEITNCFPFPQHTEDDVDFDEVQY--QMEMMRSLRHVNIDHLHVGW 101
Query 63 YHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLMLG 122
Y S +G + S +++Q S++ + +V ++ DPI++ +G + + +RL P + +
Sbjct 102 YQS-TFYGTFVSRALLDSQFSYQHAIEESVVLIYDPIKTSQGSLSLKAYRL-TPKLMEIC 159
Query 123 QE 124
+E
Sbjct 160 KE 161
> dre:445306 eif3ha, eIF3h-A, eif3h, eif3s3, fj02c04, wu:fj02c04,
zgc:101085; eukaryotic translation initiation factor 3,
subunit H, a; K03247 translation initiation factor 3 subunit
H
Length=335
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/121 (25%), Positives = 64/121 (52%), Gaps = 4/121 (3%)
Query 4 VMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGWY 63
V G++LG ++D + + + F PQ +E + YQ EM+ L+ + VGWY
Sbjct 46 VQGVLLGLVVED-RLEITNCFPFPQHTED-DVEFDEVQYQMEMMRSLRHVNIDHLHVGWY 103
Query 64 HSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLMLGQ 123
S +G + S +++Q S++ + +V ++ DP+++ +G + + +RL P + + +
Sbjct 104 QS-TYYGSFVSRALLDSQFSYQHAIEESVVLIYDPLKTAQGSLCLKAYRL-TPKLMEICK 161
Query 124 E 124
E
Sbjct 162 E 162
> dre:567151 eif3hb, MGC123049, zgc:123049; eukaryotic translation
initiation factor 3, subunit H, b; K03247 translation initiation
factor 3 subunit H
Length=333
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 65/122 (53%), Gaps = 6/122 (4%)
Query 4 VMGLMLGEFIDDFTVRVVDVFSMPQ-SGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGW 62
V G++LG ++D + + + F PQ + + + V Y Q EM+ L+ + VGW
Sbjct 44 VQGVLLGLVVED-QLEITNCFPFPQHTEDDADFDEVQY--QMEMMRSLRHVNIDHLHVGW 100
Query 63 YHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLMLG 122
Y S +G + S +++Q S++ + +V ++ DPI++ +G + + +RL P + +
Sbjct 101 YQST-LYGSFVSRALLDSQFSYQHAIEESVVLIYDPIKTAQGSLCLKAYRL-TPKLMEIC 158
Query 123 QE 124
+E
Sbjct 159 KE 160
> mmu:68135 Eif3h, 1110008A16Rik, 40kD, 9430017H16Rik, EIF3-P40,
EIF3-gamma, Eif3s3, MGC107627; eukaryotic translation initiation
factor 3, subunit H; K03247 translation initiation factor
3 subunit H
Length=352
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/122 (25%), Positives = 65/122 (53%), Gaps = 6/122 (4%)
Query 4 VMGLMLGEFIDDFTVRVVDVFSMPQ-SGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGW 62
V G++LG ++D + + + F PQ + + + V Y Q EM+ L+ + VGW
Sbjct 63 VQGVLLGLVVED-RLEITNCFPFPQHTEDDADFDEVQY--QMEMMRSLRHVNIDHLHVGW 119
Query 63 YHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLMLG 122
Y S +G + + +++Q S++ + +V ++ DPI++ +G + + +RL P + +
Sbjct 120 YQS-TYYGSFVTRALLDSQFSYQHAIEESVVLIYDPIKTAQGSLSLKAYRL-TPKLMEVC 177
Query 123 QE 124
+E
Sbjct 178 KE 179
> hsa:8667 EIF3H, EIF3S3, MGC102958, eIF3-gamma, eIF3-p40; eukaryotic
translation initiation factor 3, subunit H; K03247 translation
initiation factor 3 subunit H
Length=352
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 60/111 (54%), Gaps = 5/111 (4%)
Query 4 VMGLMLGEFIDDFTVRVVDVFSMPQ-SGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGW 62
V G++LG ++D + + + F PQ + + + V Y Q EM+ L+ + VGW
Sbjct 63 VQGVLLGLVVED-RLEITNCFPFPQHTEDDADFDEVQY--QMEMMRSLRHVNIDHLHVGW 119
Query 63 YHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRL 113
Y S +G + + +++Q S++ + +V ++ DPI++ +G + + +RL
Sbjct 120 YQS-TYYGSFVTRALLDSQFSYQHAIEESVVLIYDPIKTAQGSLSLKAYRL 169
> ath:AT1G10600 hypothetical protein
Length=166
Score = 46.6 bits (109), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINP 116
VGW H+HP GC+ S D++T S++ + A +VV P S K + F+L +P
Sbjct 66 VGWIHTHPSQGCFMSSVDLHTHYSYQVMVPEAFAIVVAPTDSSKSYGI---FKLTDP 119
> xla:779416 mysm1, 2A-DUB, MGC154322; myb-like, SWIRM and MPN
domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=818
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 55/102 (53%), Gaps = 10/102 (9%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSM-PQSGNSVSIEA-VDYVYQTEMLDQLKRTGRPEMVV 60
EV+GL+ G + + + VV++ ++ P + S ++ +D V QT+ + L G V+
Sbjct 568 EVIGLLGGRYTE--SESVVEICAVEPCNSLSTGLQCEMDPVSQTQASEALASRGY--SVI 623
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRA----VGVVVDP 98
GWYHSHP F S D++TQ ++ R +G+++ P
Sbjct 624 GWYHSHPAFDPNPSIRDIDTQAKYQNYFSRGGAKFLGMIISP 665
> hsa:114803 MYSM1, 2A-DUB, 2ADUB, DKFZp779J1554, DKFZp779J1721,
KIAA1915, RP4-592A1.1; Myb-like, SWIRM and MPN domains 1
(EC:3.1.2.15); K11865 protein MYSM1
Length=828
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 54/102 (52%), Gaps = 10/102 (9%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSM-PQSGNSVSIEA-VDYVYQTEMLDQLKRTGRPEMVV 60
EV+GL+ G + + +VV+V + P + S ++ +D V QT+ + L G V+
Sbjct 597 EVIGLLGGRYSE--VDKVVEVCAAEPCNSLSTGLQCEMDPVSQTQASETLAVRGF--SVI 652
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRA----VGVVVDP 98
GWYHSHP F S D++TQ ++ R +G++V P
Sbjct 653 GWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAKFIGMIVSP 694
> mmu:320713 Mysm1, C130067A03Rik, C530050H10Rik; myb-like, SWIRM
and MPN domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=819
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 54/102 (52%), Gaps = 10/102 (9%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSM-PQSGNSVSIEA-VDYVYQTEMLDQLKRTGRPEMVV 60
EV+GL+ G + + +V++V + P + S ++ +D V QT+ + L G V+
Sbjct 588 EVIGLLGGRYSE--ADKVLEVCAAEPCNSLSTGLQCEMDPVSQTQASETLALRGY--SVI 643
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRA----VGVVVDP 98
GWYHSHP F S D++TQ ++ R +G++V P
Sbjct 644 GWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAKFIGMIVSP 685
> ath:AT1G80210 hypothetical protein; K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=371
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 52/113 (46%), Gaps = 9/113 (7%)
Query 3 EVMGLMLG--EFIDDFTVRVVDVF-SMPQSGNSVSIEAVD-----YVYQTEMLDQLK-RT 53
E+MGL+LG E+ D ++ + PQS + + V+ + D++ T
Sbjct 27 EIMGLLLGDIEYSKDGGSATAMIWGASPQSRSDRQKDRVETNPEQLAAASAQADRMTIST 86
Query 54 GRPEMVVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKV 106
GR V+GWYHSHP S DV TQ ++ L+ +G++ KV
Sbjct 87 GRTTRVIGWYHSHPHITVLPSHVDVRTQAMYQLLDSGFIGLIFSCFSEDANKV 139
> dre:100331111 histone H2A deubiquitinase MYSM1-like
Length=2360
Score = 44.7 bits (104), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/103 (33%), Positives = 54/103 (52%), Gaps = 12/103 (11%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIE---AVDYVYQTEMLDQLKRTGRPEMV 59
EV+GL+ G + ++ +V+ + S + NS+S +D V QT+ + L G V
Sbjct 2075 EVIGLLGGTYEEED--KVLKICSA-EPCNSLSTGLQCEMDPVSQTQASEVLGVKGL--SV 2129
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRA----VGVVVDP 98
VGWYHSHP F S D++TQ ++ R +G++V P
Sbjct 2130 VGWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAPFIGMIVSP 2172
> dre:561225 mysm1, im:7153144, si:ch211-59d15.8; Myb-like, SWIRM
and MPN domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=822
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 52/101 (51%), Gaps = 8/101 (7%)
Query 3 EVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEA-VDYVYQTEMLDQLKRTGRPEMVVG 61
EV+GL+ G + ++ V + + P + S ++ +D V QT+ + L G VVG
Sbjct 537 EVIGLLGGTYEEEDKVLKI-CSAEPCNSLSTGLQCEMDPVSQTQASEVLGVKGL--SVVG 593
Query 62 WYHSHPGFGCWFSGTDVNTQQSFEQLNQRA----VGVVVDP 98
WYHSHP F S D++TQ ++ R +G++V P
Sbjct 594 WYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAPFIGMIVSP 634
> ath:AT3G06820 mov34 family protein; K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=405
Score = 44.3 bits (103), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 48/113 (42%), Gaps = 9/113 (7%)
Query 3 EVMGLMLGEFI-----DDFTVRVVDVFSMPQSG---NSVSIEAVDYVYQTEMLDQLK-RT 53
E+MGL+LG+ + T + P+S + V + D++ T
Sbjct 27 EIMGLLLGDIEYSKNGESATAMIWGASPQPRSDRQKDRVETNPEQLAAASAQADRMTIST 86
Query 54 GRPEMVVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKV 106
GR V+GWYHSHP S DV TQ ++ L+ +G++ KV
Sbjct 87 GRTTRVIGWYHSHPHITVLPSHVDVRTQAMYQLLDSGFIGLIFSCFSEDANKV 139
> ath:AT1G48790 mov34 family protein; K11866 STAM-binding protein
[EC:3.1.2.15]
Length=507
Score = 43.5 bits (101), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Query 60 VGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINP 116
+GW H+HP C+ S DV+T S++ + AV +V+ P S + + FRL P
Sbjct 407 LGWIHTHPTQSCFMSSIDVHTHYSYQIMLPEAVAIVMAPQDSSRNHGI---FRLTTP 460
> xla:444711 stambpl1, MGC84444; STAM binding protein-like 1 (EC:3.1.2.15);
K11867 AMSH-like protease [EC:3.1.2.15]
Length=431
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/141 (22%), Positives = 61/141 (43%), Gaps = 24/141 (17%)
Query 2 MEVMGLMLGEFI-DDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
+E G++ G+ D+FT+ V V + +E V+ ++ + L + +
Sbjct 286 IETCGILCGKLTHDEFTITHVIVPKQSAGPDYCDMENVEELFNVQDQHNL-------LTL 338
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GW H+HP + S D++T S++ + A+ +V P + G FRL + L
Sbjct 339 GWIHTHPTQTAFLSSVDLHTHCSYQLMLPEAIAIVCSPKHNDTG-----IFRLTSAGMLE 393
Query 121 L-----------GQEPRQTTS 130
+ +EPRQ ++
Sbjct 394 VSVCKKKGFHPHSKEPRQFST 414
> xla:414627 hypothetical protein MGC81376; K11867 AMSH-like protease
[EC:3.1.2.15]
Length=431
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/138 (23%), Positives = 59/138 (42%), Gaps = 24/138 (17%)
Query 2 MEVMGLMLGEFI-DDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
+E G++ G+ D+FT+ V V + +E V+ ++ + L + +
Sbjct 286 IETCGILCGKLTHDEFTITHVIVPKQSAGPDYCDMENVEELFNVQDQHNL-------LTL 338
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFLM 120
GW H+HP + S D++T S++ + A+ +V P + G FRL + L
Sbjct 339 GWIHTHPTQTAFLSSVDLHTHCSYQLMLPEAIAIVCSPKHNDTG-----IFRLTSAGMLE 393
Query 121 L-----------GQEPRQ 127
+ +EPRQ
Sbjct 394 VSACKKKGFHPHSKEPRQ 411
> hsa:57559 STAMBPL1, ALMalpha, AMSH-FP, AMSH-LP, FLJ31524, KIAA1373,
bA399O19.2; STAM binding protein-like 1 (EC:3.1.2.15);
K11867 AMSH-like protease [EC:3.1.2.15]
Length=436
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 52/119 (43%), Gaps = 13/119 (10%)
Query 2 MEVMGLMLGEFI-DDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
+E G++ G+ ++FT+ V V + +E V+ ++ + L + +
Sbjct 291 IETCGILCGKLTHNEFTITHVIVPKQSAGPDYCDMENVEELFNVQDQHDL-------LTL 343
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLINPHFL 119
GW H+HP + S D++T S++ + A+ +V P G FRL N L
Sbjct 344 GWIHTHPTQTAFLSSVDLHTHCSYQLMLPEAIAIVCSPKHKDTG-----IFRLTNAGML 397
> mmu:76630 Stambpl1, 1700095N21Rik, 8230401J17Rik, ALMalpha,
AMSH-FP; STAM binding protein like 1 (EC:3.1.2.15); K11867 AMSH-like
protease [EC:3.1.2.15]
Length=436
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 51/115 (44%), Gaps = 13/115 (11%)
Query 2 MEVMGLMLGEFI-DDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
+E G++ G+ ++FT+ V V + +E V+ ++ + L + +
Sbjct 291 IETCGILCGKLTHNEFTITHVVVPKQSAGPDYCDVENVEELFNVQDQHGL-------LTL 343
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRLIN 115
GW H+HP + S D++T S++ + A+ +V P G FRL N
Sbjct 344 GWIHTHPTQTAFLSSVDLHTHCSYQLMLPEAIAIVCSPKHKDTG-----IFRLTN 393
> ath:AT1G10840 TIF3H1; TIF3H1; translation initiation factor;
K03247 translation initiation factor 3 subunit H
Length=337
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 51/113 (45%), Gaps = 2/113 (1%)
Query 1 PMEVMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVV 60
P V G +LG + + V + F P + IEA YQ EM+ L+ V
Sbjct 44 PTLVTGQLLGLDVGS-VLEVTNCFPFPVRDDDEEIEADGANYQLEMMRCLREVNVDNNTV 102
Query 61 GWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPIQSVKGKVVMDCFRL 113
GWY S G + + + T ++++ +R V ++ DP ++ G + + +L
Sbjct 103 GWYQST-VLGSYQTVELIETFMNYQENIKRCVCIIYDPSKADLGVLALKALKL 154
> sce:YOR261C RPN8; Essential, non-ATPase regulatory subunit of
the 26S proteasome; has similarity to the human p40 proteasomal
subunit and to another S. cerevisiae regulatory subunit,
Rpn11p; K03038 26S proteasome regulatory subunit N8
Length=338
Score = 40.0 bits (92), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 45/87 (51%), Gaps = 7/87 (8%)
Query 5 MGLMLGEFIDDFTVRVVDVFSMP--QSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGW 62
+G++LG+ + T+RV + F++P + + + +D+ Y M + K+ E ++GW
Sbjct 34 VGVILGD-ANSSTIRVTNSFALPFEEDEKNSDVWFLDHNYIENMNEMCKKINAKEKLIGW 92
Query 63 YHSHPGFGCWFSGTDVNTQQSFEQLNQ 89
YHS P +D+ + F++ Q
Sbjct 93 YHSGPK----LRASDLKINELFKKYTQ 115
> ath:AT2G39990 EIF2; EIF2; translation initiation factor; K03249
translation initiation factor 3 subunit F
Length=293
Score = 39.7 bits (91), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Query 4 VMGLMLGEFIDDFTVRVVDVFSMPQSGNSVSIEAVDYVYQTEMLDQLKRTGRPEMVVGWY 63
V+G +LG + D TV + + +++P + +S + AVD Y ML + E +VGWY
Sbjct 52 VIGTLLGSILPDGTVDIRNSYAVPHNESSDQV-AVDIDYHHNMLASHLKVNSKETIVGWY 110
Query 64 HSHPG 68
+ G
Sbjct 111 STGAG 115
> cel:C41D11.2 eif-3.H; Eukaryotic Initiation Factor family member
(eif-3.H); K03247 translation initiation factor 3 subunit
H
Length=365
Score = 38.9 bits (89), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 41/74 (55%), Gaps = 4/74 (5%)
Query 43 QTEMLDQLKRTGRPEM---VVGWYHSHPGFGCWFSGTDVNTQQSFEQLNQRAVGVVVDPI 99
Q EMLD L++ + +VG+Y SH FG FS V + ++ + V ++ DPI
Sbjct 90 QHEMLDMLRKFRTMNIDYEIVGFYQSHQ-FGAGFSHDLVESMFDYQAMGPENVVLIYDPI 148
Query 100 QSVKGKVVMDCFRL 113
++ +G++ + +RL
Sbjct 149 KTRQGQLSLRAWRL 162
Lambda K H
0.320 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9596524200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40