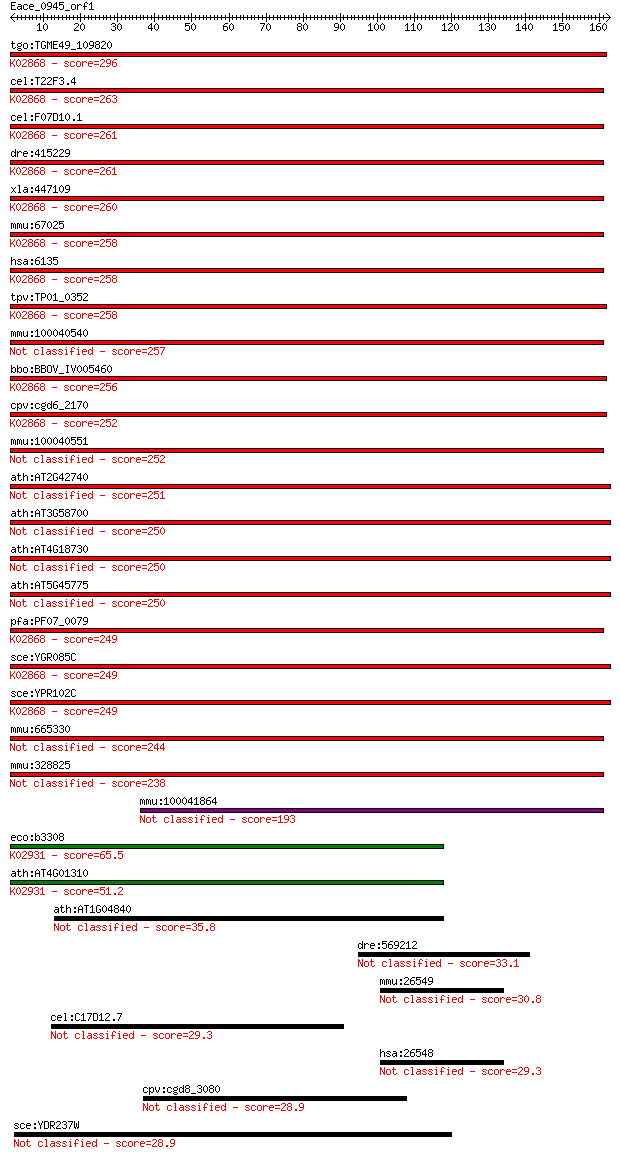
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0945_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_109820 ribosomal protein L11, putative ; K02868 lar... 296 2e-80
cel:T22F3.4 rpl-11.1; Ribosomal Protein, Large subunit family ... 263 2e-70
cel:F07D10.1 rpl-11.2; Ribosomal Protein, Large subunit family... 261 6e-70
dre:415229 rpl11, zgc:86748; ribosomal protein L11; K02868 lar... 261 1e-69
xla:447109 rpl11, MGC85310; ribosomal protein L11; K02868 larg... 260 1e-69
mmu:67025 Rpl11, 2010203J19Rik; ribosomal protein L11; K02868 ... 258 6e-69
hsa:6135 RPL11, DBA7, GIG34; ribosomal protein L11; K02868 lar... 258 6e-69
tpv:TP01_0352 60S ribosomal protein L11a; K02868 large subunit... 258 7e-69
mmu:100040540 Gm10288; ribosomal protein L11 pseudogene 257 1e-68
bbo:BBOV_IV005460 23.m05801; 60S ribosomal protein L11; K02868... 256 3e-68
cpv:cgd6_2170 60S ribosomal protein L11 ; K02868 large subunit... 252 3e-67
mmu:100040551 Gm10036; predicted gene 10036 252 4e-67
ath:AT2G42740 RPL16A; RPL16A; structural constituent of ribosome 251 7e-67
ath:AT3G58700 60S ribosomal protein L11 (RPL11B) 250 1e-66
ath:AT4G18730 RPL16B; RPL16B; structural constituent of ribosome 250 1e-66
ath:AT5G45775 60S ribosomal protein L11 (RPL11D) 250 1e-66
pfa:PF07_0079 60S ribosomal protein L11a, putative; K02868 lar... 249 2e-66
sce:YGR085C RPL11B; Rpl11bp; K02868 large subunit ribosomal pr... 249 2e-66
sce:YPR102C RPL11A; Rpl11ap; K02868 large subunit ribosomal pr... 249 2e-66
mmu:665330 Gm7589, EG665330; predicted gene 7589 244 6e-65
mmu:328825 Gm5093, EG328825; predicted gene 5093 238 5e-63
mmu:100041864 Gm3552; predicted gene 3552 193 2e-49
eco:b3308 rplE, ECK3295, JW3270; 50S ribosomal subunit protein... 65.5 7e-11
ath:AT4G01310 ribosomal protein L5 family protein; K02931 larg... 51.2 1e-06
ath:AT1G04840 pentatricopeptide (PPR) repeat-containing protein 35.8 0.064
dre:569212 tecpr2, si:dkey-46n18.5; tectonin beta-propeller re... 33.1 0.36
mmu:26549 Itgb1bp2, Chordc3; integrin beta 1 binding protein 2 30.8
cel:C17D12.7 hypothetical protein 29.3 5.6
hsa:26548 ITGB1BP2, CHORDC3, ITGB1BP, MELUSIN, MGC119214; inte... 29.3 5.9
cpv:cgd8_3080 Swr1p like SWI/SNF2 family ATpase with a HSA dom... 28.9 7.7
sce:YDR237W MRPL7; Mrpl7p 28.9 8.7
> tgo:TGME49_109820 ribosomal protein L11, putative ; K02868 large
subunit ribosomal protein L11e
Length=175
Score = 296 bits (759), Expect = 2e-80, Method: Compositional matrix adjust.
Identities = 143/161 (88%), Positives = 151/161 (93%), Gaps = 0/161 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RIEKLTLNICVGESGDRLTRAARVLEQLTGQRP S+AR TIRSF IRRNEKIAC+VTVR
Sbjct 14 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPQFSKARFTIRSFGIRRNEKIACYVTVR 73
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
GKKAE+ILEKGLKVKEYELKKKNFSD+GNFGFGIQEHIDLGIKYDPSTGIYG+DFYV L
Sbjct 74 GKKAEDILEKGLKVKEYELKKKNFSDSGNFGFGIQEHIDLGIKYDPSTGIYGMDFYVQLT 133
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLN 161
RPG RV+ RK ARGR+GHSHRVTKEDSI+WFQQ YDGIVLN
Sbjct 134 RPGNRVAHRKRARGRVGHSHRVTKEDSIKWFQQTYDGIVLN 174
> cel:T22F3.4 rpl-11.1; Ribosomal Protein, Large subunit family
member (rpl-11.1); K02868 large subunit ribosomal protein
L11e
Length=196
Score = 263 bits (671), Expect = 2e-70, Method: Compositional matrix adjust.
Identities = 127/160 (79%), Positives = 142/160 (88%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+I+KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+R+F IRRNEKIA H TVR
Sbjct 23 KIQKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRTFGIRRNEKIAVHCTVR 82
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKVKEYEL K+NFSDTGNFGFG+QEHIDLGIKYDP GIYG+DFYV L
Sbjct 83 GPKAEEILEKGLKVKEYELFKENFSDTGNFGFGVQEHIDLGIKYDPGIGIYGMDFYVVLN 142
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
R G+RVSKR+ A GRIG SHRV KE++I+WFQQKYDGI+L
Sbjct 143 RNGVRVSKRRRAPGRIGPSHRVDKEETIKWFQQKYDGIIL 182
> cel:F07D10.1 rpl-11.2; Ribosomal Protein, Large subunit family
member (rpl-11.2); K02868 large subunit ribosomal protein
L11e
Length=196
Score = 261 bits (667), Expect = 6e-70, Method: Compositional matrix adjust.
Identities = 125/160 (78%), Positives = 142/160 (88%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+I+KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+R+F IRRNEKIA H TVR
Sbjct 23 KIQKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRTFGIRRNEKIAVHCTVR 82
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKVKEYEL K+NFSDTGNFGFG+QEHIDLGIKYDPS GIYG+DFYV L
Sbjct 83 GPKAEEILEKGLKVKEYELYKENFSDTGNFGFGVQEHIDLGIKYDPSIGIYGMDFYVVLD 142
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
R G R++KR+ A GR+G SHRV +E+SI+WFQQKYDGI+L
Sbjct 143 RAGRRIAKRRRAPGRVGPSHRVEREESIKWFQQKYDGIIL 182
> dre:415229 rpl11, zgc:86748; ribosomal protein L11; K02868 large
subunit ribosomal protein L11e
Length=178
Score = 261 bits (666), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 125/160 (78%), Positives = 138/160 (86%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+RSF IRRNEKIA H TVR
Sbjct 16 RIRKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKIAVHCTVR 75
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQEHIDLGIKYDPS GIYGLDFYV L
Sbjct 76 GAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQEHIDLGIKYDPSIGIYGLDFYVVLG 135
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K GRIG HR+ KE+++RWFQQKYDGI+L
Sbjct 136 RPGFSIADKKRKTGRIGAKHRIRKEEAMRWFQQKYDGIIL 175
> xla:447109 rpl11, MGC85310; ribosomal protein L11; K02868 large
subunit ribosomal protein L11e
Length=177
Score = 260 bits (665), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 125/160 (78%), Positives = 138/160 (86%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+RSF IRRNEKIA H TVR
Sbjct 15 RIRKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKIAVHCTVR 74
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQEHIDLGIKYDPS GIYGLDFYV L
Sbjct 75 GAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQEHIDLGIKYDPSIGIYGLDFYVVLG 134
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K RG IG HR+ KE+++RWFQQKYDGI+L
Sbjct 135 RPGFSIADKKRKRGVIGSKHRIGKEEAMRWFQQKYDGIIL 174
> mmu:67025 Rpl11, 2010203J19Rik; ribosomal protein L11; K02868
large subunit ribosomal protein L11e
Length=178
Score = 258 bits (659), Expect = 6e-69, Method: Compositional matrix adjust.
Identities = 124/160 (77%), Positives = 138/160 (86%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+RSF IRRNEKIA H TVR
Sbjct 16 RIRKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKIAVHCTVR 75
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQEHIDLGIKYDPS GIYGLDFYV L
Sbjct 76 GAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQEHIDLGIKYDPSIGIYGLDFYVVLG 135
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K G IG HR++KE+++RWFQQKYDGI+L
Sbjct 136 RPGFSIADKKRRTGCIGAKHRISKEEAMRWFQQKYDGIIL 175
> hsa:6135 RPL11, DBA7, GIG34; ribosomal protein L11; K02868 large
subunit ribosomal protein L11e
Length=178
Score = 258 bits (659), Expect = 6e-69, Method: Compositional matrix adjust.
Identities = 124/160 (77%), Positives = 138/160 (86%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+RSF IRRNEKIA H TVR
Sbjct 16 RIRKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKIAVHCTVR 75
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQEHIDLGIKYDPS GIYGLDFYV L
Sbjct 76 GAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQEHIDLGIKYDPSIGIYGLDFYVVLG 135
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K G IG HR++KE+++RWFQQKYDGI+L
Sbjct 136 RPGFSIADKKRRTGCIGAKHRISKEEAMRWFQQKYDGIIL 175
> tpv:TP01_0352 60S ribosomal protein L11a; K02868 large subunit
ribosomal protein L11e
Length=171
Score = 258 bits (658), Expect = 7e-69, Method: Compositional matrix adjust.
Identities = 120/161 (74%), Positives = 140/161 (86%), Gaps = 0/161 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNI VGESGDRLTRA +VLEQLT Q+PV S+ R TIRS +RRNEKIACHVTVR
Sbjct 11 RINKLVLNIGVGESGDRLTRAGKVLEQLTDQKPVFSKCRFTIRSLGVRRNEKIACHVTVR 70
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G+KA +ILE+GLKVKEYELKKKNFSDTGNFGFGIQEHIDLG+KYDPSTGIYG+DFYV L
Sbjct 71 GQKALDILERGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGLKYDPSTGIYGMDFYVQLV 130
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLN 161
RPG RV KR+ + R+G SH+VTKE++++WFQ K+DG++ N
Sbjct 131 RPGYRVCKRRKCKTRVGKSHKVTKEEAMKWFQDKFDGLIFN 171
> mmu:100040540 Gm10288; ribosomal protein L11 pseudogene
Length=178
Score = 257 bits (656), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 123/160 (76%), Positives = 138/160 (86%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNICVGE+GDRLTRAA+VLEQLTGQ PV S+AR T+RSF IRRNEKIA H TVR
Sbjct 16 RIRKLCLNICVGENGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKIAVHCTVR 75
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQEHIDLGIKYDPS GIYGLDFYV L
Sbjct 76 GAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQEHIDLGIKYDPSIGIYGLDFYVVLG 135
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K G IG HR++KE+++RWFQQKYDGI+L
Sbjct 136 RPGFSIADKKRRTGCIGAKHRISKEEAMRWFQQKYDGIIL 175
> bbo:BBOV_IV005460 23.m05801; 60S ribosomal protein L11; K02868
large subunit ribosomal protein L11e
Length=171
Score = 256 bits (653), Expect = 3e-68, Method: Compositional matrix adjust.
Identities = 117/161 (72%), Positives = 140/161 (86%), Gaps = 0/161 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+I KL LN+ VGESGDRLTRA +VLEQLT Q+PV S+ R TIRS +RRNEKIACHVTVR
Sbjct 11 QIAKLVLNVGVGESGDRLTRAGKVLEQLTDQKPVFSKCRFTIRSLGVRRNEKIACHVTVR 70
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
GKKA E+LE+GLKVKEYELK +NFS+TGNFGFGIQEHIDLG+KYDPSTGIYG+DFYV L
Sbjct 71 GKKALELLERGLKVKEYELKSENFSNTGNFGFGIQEHIDLGLKYDPSTGIYGMDFYVQLI 130
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLN 161
RPG RV+KR+ + +IG H+VTKED+++WFQ+K+DGI+ N
Sbjct 131 RPGYRVTKRRKCKSKIGKQHKVTKEDAMKWFQEKFDGIIFN 171
> cpv:cgd6_2170 60S ribosomal protein L11 ; K02868 large subunit
ribosomal protein L11e
Length=172
Score = 252 bits (644), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 119/161 (73%), Positives = 140/161 (86%), Gaps = 0/161 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+IEKL +NI VG+SGDRLTRAA+VLEQLT Q+PV +AR TIRSFSIRR EKI+C+VTVR
Sbjct 12 KIEKLVINISVGQSGDRLTRAAKVLEQLTDQKPVFGQARFTIRSFSIRRAEKISCYVTVR 71
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKVKEYEL+K+NFS TGNFGFGI EHIDLGIKYDPSTGIYG+DF+V L
Sbjct 72 GDKAEEILEKGLKVKEYELRKRNFSATGNFGFGIDEHIDLGIKYDPSTGIYGMDFFVQLT 131
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLN 161
RPG RVS R+ R ++G RVTK+++++WFQ KYDGI+LN
Sbjct 132 RPGNRVSLRRKCRSKVGKHGRVTKDEAMQWFQSKYDGIILN 172
> mmu:100040551 Gm10036; predicted gene 10036
Length=178
Score = 252 bits (643), Expect = 4e-67, Method: Compositional matrix adjust.
Identities = 122/160 (76%), Positives = 136/160 (85%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNICVGESGDRLTRAA+VLEQLTGQ PV S+AR T+RSF I RNEKIA H TVR
Sbjct 16 RIRKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIWRNEKIAVHCTVR 75
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQEHID GIKYDPS GIYGLDFYV L
Sbjct 76 GAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQEHIDPGIKYDPSIGIYGLDFYVVLG 135
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K G IG HR++KE+++RWFQQKYDGI+L
Sbjct 136 RPGFSIADKKRRTGCIGAKHRISKEEAMRWFQQKYDGIIL 175
> ath:AT2G42740 RPL16A; RPL16A; structural constituent of ribosome
Length=182
Score = 251 bits (641), Expect = 7e-67, Method: Compositional matrix adjust.
Identities = 115/162 (70%), Positives = 143/162 (88%), Gaps = 0/162 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+++KL LNI VGESGDRLTRA++VLEQL+GQ PV S+AR T+RSF IRRNEKIAC+VTVR
Sbjct 15 KVQKLVLNISVGESGDRLTRASKVLEQLSGQTPVFSKARYTVRSFGIRRNEKIACYVTVR 74
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G+KA ++LE GLKVKEYEL ++NFSDTG FGFGIQEHIDLGIKYDPSTGIYG+DFYV L
Sbjct 75 GEKAMQLLESGLKVKEYELLRRNFSDTGCFGFGIQEHIDLGIKYDPSTGIYGMDFYVVLE 134
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLNR 162
RPG RV++R+ + R+G HRVTK+D+++WFQ KY+G++LN+
Sbjct 135 RPGYRVARRRRCKARVGIQHRVTKDDAMKWFQVKYEGVILNK 176
> ath:AT3G58700 60S ribosomal protein L11 (RPL11B)
Length=182
Score = 250 bits (639), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 115/162 (70%), Positives = 143/162 (88%), Gaps = 0/162 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+++KL LNI VGESGDRLTRA++VLEQL+GQ PV S+AR T+RSF IRRNEKIAC+VTVR
Sbjct 15 KVQKLVLNISVGESGDRLTRASKVLEQLSGQTPVFSKARYTVRSFGIRRNEKIACYVTVR 74
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G+KA ++LE GLKVKEYEL ++NFSDTG FGFGIQEHIDLGIKYDPSTGIYG+DFYV L
Sbjct 75 GEKAMQLLESGLKVKEYELLRRNFSDTGCFGFGIQEHIDLGIKYDPSTGIYGMDFYVVLE 134
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLNR 162
RPG RV++R+ + R+G HRVTK+D+++WFQ KY+G++LN+
Sbjct 135 RPGYRVARRRRCKTRVGIQHRVTKDDAMKWFQVKYEGVILNK 176
> ath:AT4G18730 RPL16B; RPL16B; structural constituent of ribosome
Length=182
Score = 250 bits (639), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 115/162 (70%), Positives = 143/162 (88%), Gaps = 0/162 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+++KL LNI VGESGDRLTRA++VLEQL+GQ PV S+AR T+RSF IRRNEKIAC+VTVR
Sbjct 15 KVQKLVLNISVGESGDRLTRASKVLEQLSGQTPVFSKARYTVRSFGIRRNEKIACYVTVR 74
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G+KA ++LE GLKVKEYEL ++NFSDTG FGFGIQEHIDLGIKYDPSTGIYG+DFYV L
Sbjct 75 GEKAMQLLESGLKVKEYELLRRNFSDTGCFGFGIQEHIDLGIKYDPSTGIYGMDFYVVLE 134
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLNR 162
RPG RV++R+ + R+G HRVTK+D+++WFQ KY+G++LN+
Sbjct 135 RPGYRVARRRRCKTRVGIQHRVTKDDAMKWFQVKYEGVILNK 176
> ath:AT5G45775 60S ribosomal protein L11 (RPL11D)
Length=182
Score = 250 bits (639), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 115/162 (70%), Positives = 143/162 (88%), Gaps = 0/162 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+++KL LNI VGESGDRLTRA++VLEQL+GQ PV S+AR T+RSF IRRNEKIAC+VTVR
Sbjct 15 KVQKLVLNISVGESGDRLTRASKVLEQLSGQTPVFSKARYTVRSFGIRRNEKIACYVTVR 74
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G+KA ++LE GLKVKEYEL ++NFSDTG FGFGIQEHIDLGIKYDPSTGIYG+DFYV L
Sbjct 75 GEKAMQLLESGLKVKEYELLRRNFSDTGCFGFGIQEHIDLGIKYDPSTGIYGMDFYVVLE 134
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLNR 162
RPG RV++R+ + R+G HRVTK+D+++WFQ KY+G++LN+
Sbjct 135 RPGYRVARRRRCKTRVGIQHRVTKDDAMKWFQVKYEGVILNK 176
> pfa:PF07_0079 60S ribosomal protein L11a, putative; K02868 large
subunit ribosomal protein L11e
Length=173
Score = 249 bits (637), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 120/160 (75%), Positives = 140/160 (87%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
++ KL LNICVGESGDRLTRAARVLEQLT Q+P+ + R TIRSF +RRNEKI+C VTVR
Sbjct 13 KVNKLVLNICVGESGDRLTRAARVLEQLTEQKPIFGKCRFTIRSFGVRRNEKISCFVTVR 72
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
GKKA EILEKGLKVKEYEL++KNFSDTGNFGFGIQEHIDLGIKYDPSTGIYG+DFYVHL
Sbjct 73 GKKALEILEKGLKVKEYELRRKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGMDFYVHLS 132
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
R G RV++R R +I +H+VTKED+++WFQ K+DGI+L
Sbjct 133 RSGYRVTRRTRRRSKISKTHKVTKEDAMKWFQTKFDGILL 172
> sce:YGR085C RPL11B; Rpl11bp; K02868 large subunit ribosomal
protein L11e
Length=174
Score = 249 bits (636), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 115/162 (70%), Positives = 140/162 (86%), Gaps = 0/162 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+IEKL LNI VGESGDRLTRA++VLEQL+GQ PV S+AR T+R+F IRRNEKIA HVTVR
Sbjct 13 KIEKLVLNISVGESGDRLTRASKVLEQLSGQTPVQSKARYTVRTFGIRRNEKIAVHVTVR 72
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILE+GLKVKEY+L+ +NFS TGNFGFGI EHIDLGIKYDPS GI+G+DFYV +
Sbjct 73 GPKAEEILERGLKVKEYQLRDRNFSATGNFGFGIDEHIDLGIKYDPSIGIFGMDFYVVMN 132
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLNR 162
RPG RV++RK +G +G+SH+ TKED++ WF+QKYD VL++
Sbjct 133 RPGARVTRRKRCKGTVGNSHKTTKEDTVSWFKQKYDADVLDK 174
> sce:YPR102C RPL11A; Rpl11ap; K02868 large subunit ribosomal
protein L11e
Length=174
Score = 249 bits (636), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 115/162 (70%), Positives = 140/162 (86%), Gaps = 0/162 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
+IEKL LNI VGESGDRLTRA++VLEQL+GQ PV S+AR T+R+F IRRNEKIA HVTVR
Sbjct 13 KIEKLVLNISVGESGDRLTRASKVLEQLSGQTPVQSKARYTVRTFGIRRNEKIAVHVTVR 72
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILE+GLKVKEY+L+ +NFS TGNFGFGI EHIDLGIKYDPS GI+G+DFYV +
Sbjct 73 GPKAEEILERGLKVKEYQLRDRNFSATGNFGFGIDEHIDLGIKYDPSIGIFGMDFYVVMN 132
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVLNR 162
RPG RV++RK +G +G+SH+ TKED++ WF+QKYD VL++
Sbjct 133 RPGARVTRRKRCKGTVGNSHKTTKEDTVSWFKQKYDADVLDK 174
> mmu:665330 Gm7589, EG665330; predicted gene 7589
Length=178
Score = 244 bits (624), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 119/160 (74%), Positives = 135/160 (84%), Gaps = 0/160 (0%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNIC+GES DRLTRAA+VLEQLTGQ V S+AR T+RSF IRRNEKIA H TVR
Sbjct 16 RIRKLCLNICIGESRDRLTRAAKVLEQLTGQTLVFSKARYTVRSFGIRRNEKIAVHCTVR 75
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+E EL+K FSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYV L
Sbjct 76 GAKAEEILEKGLKVRECELRKNIFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVVLG 135
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K G +G HR++KE+++RWFQQKYDGI+L
Sbjct 136 RPGFSIADKKHRTGCVGAKHRISKEEAMRWFQQKYDGIIL 175
> mmu:328825 Gm5093, EG328825; predicted gene 5093
Length=198
Score = 238 bits (608), Expect = 5e-63, Method: Compositional matrix adjust.
Identities = 118/160 (73%), Positives = 131/160 (81%), Gaps = 4/160 (2%)
Query 1 RIEKLTLNICVGESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVR 60
RI KL LNIC GDRLTRAA+VLEQLTGQ PV S+AR T+RSF IRRNEKIA H TVR
Sbjct 16 RIRKLCLNIC----GDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKIAVHCTVR 71
Query 61 GKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQEHIDLGIKYDPSTGIYGLDFYVHLC 120
G KAEEILEKGLKV+EYEL K NFSDTGNFGFG+QEHIDLGIKYDPS GIYGLDFY L
Sbjct 72 GAKAEEILEKGLKVREYELWKNNFSDTGNFGFGLQEHIDLGIKYDPSIGIYGLDFYEVLG 131
Query 121 RPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKYDGIVL 160
RPG ++ +K G IG HR++KE++ RWFQQKYDGI+L
Sbjct 132 RPGFSIADKKRRTGCIGAKHRISKEEATRWFQQKYDGIIL 171
> mmu:100041864 Gm3552; predicted gene 3552
Length=221
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 92/125 (73%), Positives = 105/125 (84%), Gaps = 0/125 (0%)
Query 36 SRARLTIRSFSIRRNEKIACHVTVRGKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGIQ 95
+RAR T+RSF IRRNEKIA H TVRG KAEEILEKGLKV+EYEL+K NFSDTGNFGFGIQ
Sbjct 67 TRARYTVRSFGIRRNEKIAVHCTVRGAKAEEILEKGLKVREYELRKNNFSDTGNFGFGIQ 126
Query 96 EHIDLGIKYDPSTGIYGLDFYVHLCRPGLRVSKRKWARGRIGHSHRVTKEDSIRWFQQKY 155
EHIDLGIKYDPS GIYGLDFYV L R G ++ +K G IG HR++KE+++RWFQQKY
Sbjct 127 EHIDLGIKYDPSIGIYGLDFYVVLGRSGFSIADKKRRTGCIGAKHRISKEEAMRWFQQKY 186
Query 156 DGIVL 160
GI+L
Sbjct 187 AGIIL 191
> eco:b3308 rplE, ECK3295, JW3270; 50S ribosomal subunit protein
L5; K02931 large subunit ribosomal protein L5
Length=179
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 75/127 (59%), Gaps = 10/127 (7%)
Query 1 RIEKLTLNICVGES-GDR--LTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHV 57
R+EK+TLN+ VGE+ D+ L AA L ++GQ+P++++AR ++ F IR+ I C V
Sbjct 30 RVEKITLNMGVGEAIADKKLLDNAAADLAAISGQKPLITKARKSVAGFKIRQGYPIGCKV 89
Query 58 TVRGKKAEEILEKGL-----KVKEYE-LKKKNFSDTGNFGFGIQEHIDLG-IKYDPSTGI 110
T+RG++ E E+ + +++++ L K+F GN+ G++E I I YD +
Sbjct 90 TLRGERMWEFFERLITIAVPRIRDFRGLSAKSFDGRGNYSMGVREQIIFPEIDYDKVDRV 149
Query 111 YGLDFYV 117
GLD +
Sbjct 150 RGLDITI 156
> ath:AT4G01310 ribosomal protein L5 family protein; K02931 large
subunit ribosomal protein L5
Length=262
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/127 (22%), Positives = 68/127 (53%), Gaps = 10/127 (7%)
Query 1 RIEKLTLNICVGESGDR---LTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHV 57
+++K+ +N +G++ L A + + +TGQ+P+ +RAR +I +F IR ++ + V
Sbjct 85 KVQKIVVNCGIGDAAQNDKGLEAAMKDIALITGQKPIKTRARASIATFKIREDQPLGIAV 144
Query 58 TVRGKKAEEILEKGL-----KVKEYE-LKKKNFSDTGNFGFGIQEH-IDLGIKYDPSTGI 110
T+RG L++ + + ++++ + +F GN+ G+++ + I++D
Sbjct 145 TLRGDVMYSFLDRLINLALPRTRDFQGVSPSSFDGNGNYSIGVKDQGVFPEIRFDAVGKT 204
Query 111 YGLDFYV 117
G+D +
Sbjct 205 RGMDVCI 211
> ath:AT1G04840 pentatricopeptide (PPR) repeat-containing protein
Length=665
Score = 35.8 bits (81), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 37/110 (33%), Positives = 53/110 (48%), Gaps = 14/110 (12%)
Query 13 ESGDRLTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVTVRGKKAEEILEKGL 72
+SG+ L RA ++ E L ++ V+S L I FS + + A E+LEKGL
Sbjct 239 DSGE-LNRAKQLFE-LMPEKNVVSWTTL-INGFSQTGDYETAISTYF------EMLEKGL 289
Query 73 KVKEYELKK--KNFSDTGNFGFGIQEH---IDLGIKYDPSTGIYGLDFYV 117
K EY + S +G G GI+ H +D GIK D + G +D Y
Sbjct 290 KPNEYTIAAVLSACSKSGALGSGIRIHGYILDNGIKLDRAIGTALVDMYA 339
> dre:569212 tecpr2, si:dkey-46n18.5; tectonin beta-propeller
repeat containing 2
Length=1308
Score = 33.1 bits (74), Expect = 0.36, Method: Composition-based stats.
Identities = 22/56 (39%), Positives = 26/56 (46%), Gaps = 11/56 (19%)
Query 95 QEHIDLGIKYDPSTGIYGLDFYVHLC----------RPGLRVSKRKWARGRIGHSH 140
Q H LG K S G +G F LC RPGLR+ R RGR+G +H
Sbjct 201 QSHQQLGSKPRKSNGKFGACFQPALCKQSDLVVYAARPGLRLW-RTDVRGRVGETH 255
> mmu:26549 Itgb1bp2, Chordc3; integrin beta 1 binding protein
2
Length=350
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 101 GIKYDPSTGIYGLDFYVHLCRPGLRVSKRKWAR 133
G+K GI LDF L +PG RV + WA+
Sbjct 181 GMKSWSCCGIQTLDFGAFLAQPGCRVGRHDWAK 213
> cel:C17D12.7 hypothetical protein
Length=657
Score = 29.3 bits (64), Expect = 5.6, Method: Composition-based stats.
Identities = 22/91 (24%), Positives = 38/91 (41%), Gaps = 12/91 (13%)
Query 12 GESGDRLTRAARVLEQLTGQRPVLSRARL------------TIRSFSIRRNEKIACHVTV 59
ES R + Q+ G +PV R + T + I +I +
Sbjct 436 AESNLRHFGGKTISAQIHGSKPVQRRPTVPKASKTGSDRPTTASTAKIVEKPEIPVEIPQ 495
Query 60 RGKKAEEILEKGLKVKEYELKKKNFSDTGNF 90
+ + + +I EK LK+ ++ +K NFS+ NF
Sbjct 496 KLEISRKIAEKTLKLDDFPVKSSNFSNISNF 526
> hsa:26548 ITGB1BP2, CHORDC3, ITGB1BP, MELUSIN, MGC119214; integrin
beta 1 binding protein (melusin) 2
Length=347
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 101 GIKYDPSTGIYGLDFYVHLCRPGLRVSKRKWAR 133
G+K GI LDF L +PG RV + W +
Sbjct 180 GMKSWSCCGIQTLDFGAFLAQPGCRVGRHDWGK 212
> cpv:cgd8_3080 Swr1p like SWI/SNF2 family ATpase with a HSA domain
at the N-terminus probably involved in chromatin remodelling
Length=1371
Score = 28.9 bits (63), Expect = 7.7, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 6/73 (8%)
Query 37 RARLTIRSFSIRRNEKIACHVT--VRGKKAEEILEKGLKVKEYELKKKNFSDTGNFGFGI 94
R R+ ++ S+ N K +++ VR +K E L K L++K++E K S+T F FG
Sbjct 79 RLRIVSKNISVNIN-KFWNNISKIVRHRKLSE-LNKLLRIKQFEKLDKLVSETEKFYFGT 136
Query 95 QEHIDLGIKYDPS 107
E D G K P+
Sbjct 137 NE--DSGKKTQPN 147
> sce:YDR237W MRPL7; Mrpl7p
Length=292
Score = 28.9 bits (63), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 26/128 (20%), Positives = 61/128 (47%), Gaps = 10/128 (7%)
Query 2 IEKLTLNICVGESGDR---LTRAARVLEQLTGQRPVLSRARLTIRSFSIRRNEKIACHVT 58
+E + +N V E+ + AA L+Q+TG +P ++ + ++ +R+ ++ V
Sbjct 137 LESVVINCFVREARENQLLAITAALQLQQITGCKPHPIFSKNDVPTWKLRKGHQMGAKVE 196
Query 59 VRGKKAEEILEKGL-----KVKEYE-LKKKNFSDTGNFGFGI-QEHIDLGIKYDPSTGIY 111
++GK+ + L +++EY+ + ++ + G FG+ E I + D + +
Sbjct 197 LKGKEMSQFLSTLTEIVLPRIREYKGISNQSGNRFGGISFGLTAEDIKFFPEIDANQDSW 256
Query 112 GLDFYVHL 119
F +H+
Sbjct 257 PKTFGMHI 264
Lambda K H
0.322 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40