bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0954_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
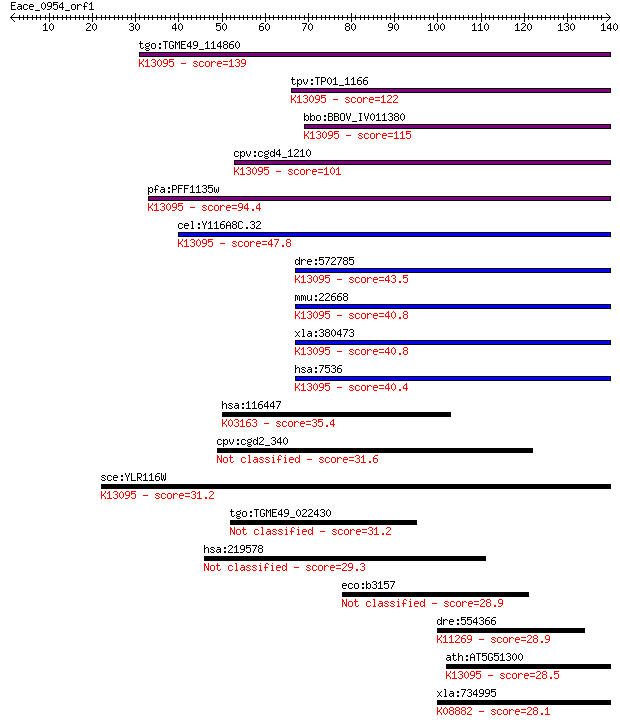
tgo:TGME49_114860 zinc knuckle domain-containing protein (EC:2... 139 3e-33
tpv:TP01_1166 transcription factor; K13095 splicing factor 1 122 3e-28
bbo:BBOV_IV011380 23.m06391; transcription or splicing factor-... 115 3e-26
cpv:cgd4_1210 Ms15p; KH + 2 Znknuckle (C2HC) ; K13095 splicing... 101 7e-22
pfa:PFF1135w transcription or splicing factor-like protein, pu... 94.4 1e-19
cel:Y116A8C.32 sfa-1; Splicing FActor family member (sfa-1); K... 47.8 1e-05
dre:572785 sf1, wu:fc09f06, znf162; splicing factor 1; K13095 ... 43.5 2e-04
mmu:22668 Sf1, BB094781, CW17R, MZFM, WBP4, Zfp162; splicing f... 40.8 0.001
xla:380473 sf1, MGC130789, MGC53716; splicing factor 1; K13095... 40.8 0.001
hsa:7536 SF1, BBP, D11S636, MBBP, ZFM1, ZNF162; splicing facto... 40.4 0.002
hsa:116447 TOP1MT; topoisomerase (DNA) I, mitochondrial (EC:5.... 35.4 0.050
cpv:cgd2_340 signal peptide, large protein 31.6 0.77
sce:YLR116W MSL5; BBP; K13095 splicing factor 1 31.2
tgo:TGME49_022430 ubiquitin-transferase domain containing prot... 31.2 1.2
hsa:219578 ZNF804B, FLJ32110; zinc finger protein 804B 29.3 4.2
eco:b3157 yhbT, ECK3145, JW3126; predicted lipid carrier prote... 28.9 4.6
dre:554366 chtf18, MGC113153, MGC192547, zgc:113153; CTF18, ch... 28.9 4.8
ath:AT5G51300 splicing factor-related; K13095 splicing factor 1 28.5
xla:734995 trim28, MGC130965, kap1, rnf96, tf1b, tif1b; tripar... 28.1 7.8
> tgo:TGME49_114860 zinc knuckle domain-containing protein (EC:2.7.7.8);
K13095 splicing factor 1
Length=723
Score = 139 bits (350), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 79/112 (70%), Positives = 91/112 (81%), Gaps = 4/112 (3%)
Query 31 MSLEQLMALVPLEAAMTHKEKG-RKSRWE--KSKKSADGSRWGSETDKPFLPPPFIDLPI 87
MS+EQLMALVPLEAA+ KEKG RKSRWE K+KK D S+WG DK FLPPP++DLP+
Sbjct 1 MSMEQLMALVPLEAALAGKEKGGRKSRWERPKTKKKGD-SKWGPPEDKDFLPPPYVDLPV 59
Query 88 GMTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
GMT Q+DRFLREQR ++L RKL NG EF D DIRPPSPPPVYD+NGGR+N
Sbjct 60 GMTAPQMDRFLREQRFDDLQRKLANGEFEFGDPDIRPPSPPPVYDRNGGRIN 111
> tpv:TP01_1166 transcription factor; K13095 splicing factor 1
Length=484
Score = 122 bits (307), Expect = 3e-28, Method: Composition-based stats.
Identities = 54/74 (72%), Positives = 63/74 (85%), Gaps = 0/74 (0%)
Query 66 GSRWGSETDKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRPP 125
G RWG E DKPFLPPP++DLP G+TP Q+D+FLREQR +EL RK+ +G LEF DA+IRPP
Sbjct 53 GCRWGPEEDKPFLPPPYVDLPPGLTPAQIDQFLREQRHDELARKITSGELEFVDAEIRPP 112
Query 126 SPPPVYDKNGGRVN 139
SPPPVYDKNG RVN
Sbjct 113 SPPPVYDKNGSRVN 126
> bbo:BBOV_IV011380 23.m06391; transcription or splicing factor-like
protein; K13095 splicing factor 1
Length=488
Score = 115 bits (289), Expect = 3e-26, Method: Composition-based stats.
Identities = 48/71 (67%), Positives = 61/71 (85%), Gaps = 0/71 (0%)
Query 69 WGSETDKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPP 128
WG E D+P+LPPP++DLP G+TP Q+D+FLREQR ++L +K+ +G LEF DADIRPPSPP
Sbjct 66 WGPEDDRPYLPPPYVDLPPGLTPSQMDQFLREQRHDDLVKKIASGELEFGDADIRPPSPP 125
Query 129 PVYDKNGGRVN 139
PVYD+NG RVN
Sbjct 126 PVYDRNGSRVN 136
> cpv:cgd4_1210 Ms15p; KH + 2 Znknuckle (C2HC) ; K13095 splicing
factor 1
Length=471
Score = 101 bits (252), Expect = 7e-22, Method: Composition-based stats.
Identities = 45/87 (51%), Positives = 60/87 (68%), Gaps = 1/87 (1%)
Query 53 RKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNN 112
R+SRW K + SRW S +K ++PP + D P GM+ ++D+FLREQRL+EL KL
Sbjct 40 RESRWSKVSDGST-SRWKSVYEKEYIPPAYSDFPPGMSNYEIDQFLREQRLDELIYKLQM 98
Query 113 GILEFFDADIRPPSPPPVYDKNGGRVN 139
G +E+ DIR PSPPP+YDKNG R+N
Sbjct 99 GEIEYGSPDIREPSPPPIYDKNGSRIN 125
> pfa:PFF1135w transcription or splicing factor-like protein,
putative; K13095 splicing factor 1
Length=615
Score = 94.4 bits (233), Expect = 1e-19, Method: Composition-based stats.
Identities = 58/114 (50%), Positives = 80/114 (70%), Gaps = 8/114 (7%)
Query 33 LEQLMALVPLEAAMTHKEKGRKSRWEKS-------KKSADGSRWGSETDKPFLPPPFIDL 85
L L+ ++P+E ++ + +KSRWE++ K ++WGSE KP+LP PF+D
Sbjct 6 LVALVNILPVEK-LSDENSKKKSRWERNATNNIIENKVVVSNKWGSEDYKPYLPLPFVDF 64
Query 86 PIGMTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
P G+TP Q+D+FLREQR +EL +KLN G LE+ D DIRPPSPPP+YDKNG R+N
Sbjct 65 PPGLTPAQLDQFLREQRYDELTKKLNKGELEYVDPDIRPPSPPPIYDKNGSRIN 118
> cel:Y116A8C.32 sfa-1; Splicing FActor family member (sfa-1);
K13095 splicing factor 1
Length=699
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 35/107 (32%), Positives = 48/107 (44%), Gaps = 21/107 (19%)
Query 40 VPLEA-------AMTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPL 92
VP+EA A+ +K R+SRW T K F+P LP +T
Sbjct 170 VPVEAVQAGAVVALNPAKKERRSRWS--------------TTKSFVPGMPTILPADLTED 215
Query 93 QVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
Q + +L + +E+ RKL + R PSP PVYD NG R+N
Sbjct 216 QRNAYLLQLEIEDATRKLRLADFGVAEGRERSPSPEPVYDANGKRLN 262
> dre:572785 sf1, wu:fc09f06, znf162; splicing factor 1; K13095
splicing factor 1
Length=565
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 42/76 (55%), Gaps = 3/76 (3%)
Query 67 SRWGSET--DKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEF-FDADIR 123
SRW SET K +P +P G+T Q ++ + ++E+L RKL G L + + R
Sbjct 20 SRWSSETPDQKTVIPGMPTVIPPGLTRDQERAYIVQLQIEDLTRKLRTGDLGIPVNPEDR 79
Query 124 PPSPPPVYDKNGGRVN 139
PSP P+Y+ G R+N
Sbjct 80 SPSPEPIYNSEGKRLN 95
> mmu:22668 Sf1, BB094781, CW17R, MZFM, WBP4, Zfp162; splicing
factor 1; K13095 splicing factor 1
Length=639
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 67 SRWGSET--DKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFF-DADIR 123
SRW +T K +P +P G+T Q ++ + ++E+L RKL G L + + R
Sbjct 20 SRWNQDTMEQKTVIPGMPTVIPPGLTREQERAYIVQLQIEDLTRKLRTGDLGIPPNPEDR 79
Query 124 PPSPPPVYDKNGGRVN 139
PSP P+Y+ G R+N
Sbjct 80 SPSPEPIYNSEGKRLN 95
> xla:380473 sf1, MGC130789, MGC53716; splicing factor 1; K13095
splicing factor 1
Length=571
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 67 SRWGSET--DKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFF-DADIR 123
SRW ET K +P +P G++ Q ++ + ++E+L RKL G L + + R
Sbjct 10 SRWNDETPDQKTIIPGMPTVIPPGLSRDQERAYIVQLQIEDLTRKLRTGDLGIPPNPEDR 69
Query 124 PPSPPPVYDKNGGRVN 139
PSP P+Y+ G R+N
Sbjct 70 SPSPEPIYNSEGKRLN 85
> hsa:7536 SF1, BBP, D11S636, MBBP, ZFM1, ZNF162; splicing factor
1; K13095 splicing factor 1
Length=673
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 67 SRWGSET--DKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFF-DADIR 123
SRW +T K +P +P G+T Q ++ + ++E+L RKL G L + + R
Sbjct 145 SRWNQDTMEQKTVIPGMPTVIPPGLTREQERAYIVQLQIEDLTRKLRTGDLGIPPNPEDR 204
Query 124 PPSPPPVYDKNGGRVN 139
PSP P+Y+ G R+N
Sbjct 205 SPSPEPIYNSEGKRLN 220
> hsa:116447 TOP1MT; topoisomerase (DNA) I, mitochondrial (EC:5.99.1.2);
K03163 DNA topoisomerase I [EC:5.99.1.2]
Length=601
Score = 35.4 bits (80), Expect = 0.050, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 8/54 (14%)
Query 50 EKGRKSRWEKSKKSADGSRWGS-ETDKPFLPPPFIDLPIGMTPLQVDRFLREQR 102
+KG +RWEK +K DG +W E P+ PP+ LP G+ RF E R
Sbjct 35 QKGSGARWEK-EKHEDGVKWRQLEHKGPYFAPPYEPLPDGV------RFFYEGR 81
> cpv:cgd2_340 signal peptide, large protein
Length=1236
Score = 31.6 bits (70), Expect = 0.77, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 37/77 (48%), Gaps = 8/77 (10%)
Query 49 KEKGRKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPLQVDRFLREQRLEELYR 108
+++G+KS+ E DG W E +L PFID + + Q+ FLR + LE +
Sbjct 598 RDQGKKSKTEFEYFGKDGRLWSVEK---YLSEPFIDRVLKLNG-QLSPFLRAKILERIKL 653
Query 109 KLNNGI----LEFFDAD 121
L I L +F+ D
Sbjct 654 TLQAFIKVFELSYFNID 670
> sce:YLR116W MSL5; BBP; K13095 splicing factor 1
Length=476
Score = 31.2 bits (69), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 46/118 (38%), Gaps = 17/118 (14%)
Query 22 SFLFNSFNKMSLEQLMALVPLEAAMTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPPP 81
S F + S+E+ A VP + S W K+ +D R+ S K
Sbjct 8 SRYFENRKGSSMEEKKAKVPPNVNL--------SLWRKNTVESDVHRFNSLPSK------ 53
Query 82 FIDLPIGMTPLQVDRFLREQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
+ +T Q+ + R++E+ KL R PSPPPVYD G R N
Sbjct 54 ---ISGALTREQIYSYQVMFRIQEITIKLRTNDFVPPSRKNRSPSPPPVYDAQGKRTN 108
> tgo:TGME49_022430 ubiquitin-transferase domain containing protein
(EC:4.2.1.1)
Length=1629
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 3/43 (6%)
Query 52 GRKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPLQV 94
G E SK +AD +R + D L PP LP TP++V
Sbjct 626 GETEHMEASKNAADAARCETAGDAAILHPP---LPYSATPVEV 665
> hsa:219578 ZNF804B, FLJ32110; zinc finger protein 804B
Length=1349
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 16/67 (23%), Positives = 33/67 (49%), Gaps = 2/67 (2%)
Query 46 MTHKEKGRKSRWEKSKKSADGSRWGSETDKPFLPPPFIDLPIGMTPLQVDRF--LREQRL 103
+ H E S ++++ + D G E + PP I PI +P ++D++ L+ Q
Sbjct 1107 INHVEGNINSYYDRTMQKPDKVEDGLEMCHKSISPPLIQQPITFSPDEIDKYKILQLQAQ 1166
Query 104 EELYRKL 110
+ + ++L
Sbjct 1167 QHMQKQL 1173
> eco:b3157 yhbT, ECK3145, JW3126; predicted lipid carrier protein,
COG3154 family
Length=174
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query 78 LPPPFIDLPIGMTPLQVDRFLREQRLEELYRK-LNNGILEFFDA 120
L P + +P+ +TP + R + EQ L +R+ L++G LEF +
Sbjct 12 LGPSLLSVPVKLTPFALKRQVLEQVLSWQFRQALDDGELEFLEG 55
> dre:554366 chtf18, MGC113153, MGC192547, zgc:113153; CTF18,
chromosome transmission fidelity factor 18 homolog (S. cerevisiae);
K11269 chromosome transmission fidelity protein 18
Length=957
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Query 100 EQRLEELYRKLNNGILEFFDADIRPPSPPPVYDK 133
++R +++ +KL G+ + D DI PPS P VYD+
Sbjct 90 KKRRQDVAKKLQFGVDQ--DDDITPPSSPEVYDR 121
> ath:AT5G51300 splicing factor-related; K13095 splicing factor
1
Length=804
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 102 RLEELYRKLNNGI-LEFFDADIRPPSPPPVYDKNGGRVN 139
RL E+ R L +G+ L+ R PSP PVYD G R+N
Sbjct 164 RLLEISRMLQSGMPLDDRPEGQRSPSPEPVYDNMGIRIN 202
> xla:734995 trim28, MGC130965, kap1, rnf96, tf1b, tif1b; tripartite
motif containing 28; K08882 tripartite motif-containing
protein 28
Length=677
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 100 EQRLEELYRKLNNGILEFFDADIRPPSPPPVYDKNGGRVN 139
E + EL +K+ LE D D+ P + PPV+ G N
Sbjct 350 EGGVNELLKKVPRVSLERLDVDLNPDTQPPVFKVFPGNTN 389
Lambda K H
0.323 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40