bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0960_orf1
Length=223
Score E
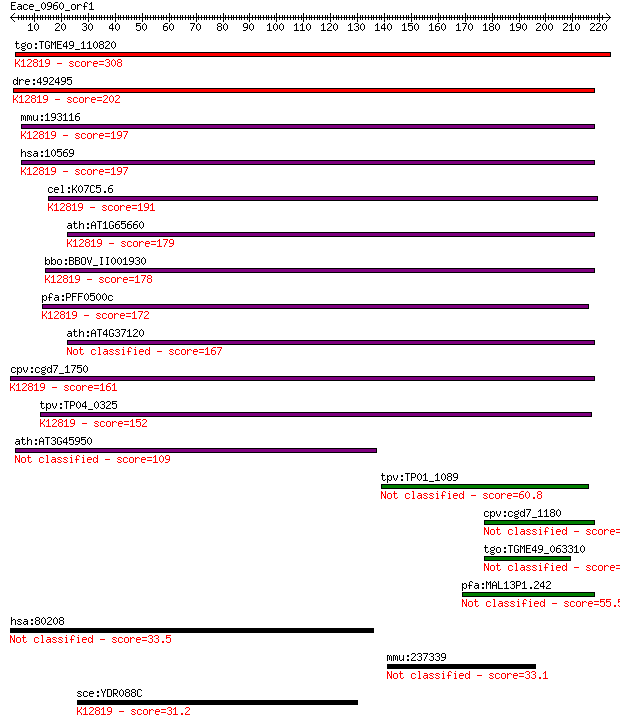
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_110820 step II splicing factor SLU7, putative ; K12... 308 1e-83
dre:492495 slu7, wu:fc94e11, zgc:103640; SLU7 splicing factor ... 202 1e-51
mmu:193116 Slu7, AU018913, D11Ertd730e, D3Bwg0878e, MGC31026; ... 197 2e-50
hsa:10569 SLU7, 9G8, MGC9280, hSlu7; SLU7 splicing factor homo... 197 3e-50
cel:K07C5.6 hypothetical protein; K12819 pre-mRNA-processing f... 191 1e-48
ath:AT1G65660 SMP1; SMP1 (SWELLMAP 1); nucleic acid binding / ... 179 9e-45
bbo:BBOV_II001930 18.m06149; mRNA processing-related protein; ... 178 1e-44
pfa:PFF0500c step II splicing factor, putative; K12819 pre-mRN... 172 8e-43
ath:AT4G37120 SMP2; SMP2; single-stranded RNA binding 167 4e-41
cpv:cgd7_1750 step II splicing factor SLU7 ; K12819 pre-mRNA-p... 161 1e-39
tpv:TP04_0325 step II splicing factor; K12819 pre-mRNA-process... 152 7e-37
ath:AT3G45950 splicing factor-related 109 6e-24
tpv:TP01_1089 hypothetical protein 60.8 3e-09
cpv:cgd7_1180 hypothetical protein 58.5 2e-08
tgo:TGME49_063310 hypothetical protein 55.8 1e-07
pfa:MAL13P1.242 step II splicing factor, putative 55.5 2e-07
hsa:80208 SPG11, DKFZp762B1512, FLJ21439, KIAA1840; spastic pa... 33.5 0.61
mmu:237339 L3mbtl3, AI481284, MBT-1, MGC90546; l(3)mbt-like 3 ... 33.1 0.89
sce:YDR088C SLU7, SLT17; RNA splicing factor, required for ATP... 31.2 2.8
> tgo:TGME49_110820 step II splicing factor SLU7, putative ; K12819
pre-mRNA-processing factor SLU7
Length=544
Score = 308 bits (788), Expect = 1e-83, Method: Compositional matrix adjust.
Identities = 146/221 (66%), Positives = 178/221 (80%), Gaps = 0/221 (0%)
Query 3 EFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQ 62
+FD+TSAPV C+D+R RINT++LRIREDTAKYLLNLD+NSAFYDPKSRSMR +PF+HL +
Sbjct 281 DFDQTSAPVACSDDRFRINTKNLRIREDTAKYLLNLDINSAFYDPKSRSMRGNPFEHLKE 340
Query 63 QIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQ 122
+ + F+GDN + G+V QQLQLFAWEAYKHG VHFNAQPTQLE LY+EH +K +
Sbjct 341 EEQALFKGDNCARKTGDVLKAQQLQLFAWEAYKHGAQVHFNAQPTQLEKLYQEHVDRKKE 400
Query 123 IETEKQKNIFDTYGGKEHLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVL 182
+E EK+ + + YGGKEHL AD R+L AQTE YVEYS+DG I +GR R L KSKYEED
Sbjct 401 LEEEKKNALLNKYGGKEHLNADPRMLLAQTEVYVEYSRDGNIAKGRNRVLIKSKYEEDAY 460
Query 183 QGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTGRLEDNI 223
G+HTSV+GS+Y++AT KWG+ CCR T FAA+CTGR ED I
Sbjct 461 VGNHTSVFGSWYNLATQKWGFKCCRQTDFAADCTGRQEDAI 501
> dre:492495 slu7, wu:fc94e11, zgc:103640; SLU7 splicing factor
homolog (S. cerevisiae); K12819 pre-mRNA-processing factor
SLU7
Length=571
Score = 202 bits (513), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 100/218 (45%), Positives = 141/218 (64%), Gaps = 4/218 (1%)
Query 2 KEFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLS 61
K D P + D + RI R+LRIRED AKYL NLD NSA+YDPK+R+MRE+P+ +
Sbjct 227 KYVDDFDMPGQNFDSKRRITVRNLRIREDIAKYLRNLDPNSAYYDPKTRAMRENPYSNTG 286
Query 62 QQIKPT-FRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKK 120
+ + + GDN + +G+ + Q QLFAWEAY+ G VH A PT+LE L++ ++ KK
Sbjct 287 KNPEEVGYAGDNFVRYSGDTISMAQTQLFAWEAYEKGSEVHLQADPTKLELLHQSYKVKK 346
Query 121 AQIETEKQKNIFDTYGGKEHLVADTR-VLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEE 179
+ ++++ I + YGG EHL A R +L AQTE YVEYS+ G +L+G+++ + +SKYEE
Sbjct 347 DDFKEKQKETILEKYGGSEHLDAPPRELLLAQTEEYVEYSRHGAVLKGQEKAVAQSKYEE 406
Query 180 DVLQGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
DVL +HT +WGSY+ WGY CC + CTG
Sbjct 407 DVLNNNHTCIWGSYW--KDGYWGYKCCHSMVKQSYCTG 442
> mmu:193116 Slu7, AU018913, D11Ertd730e, D3Bwg0878e, MGC31026;
SLU7 splicing factor homolog (S. cerevisiae); K12819 pre-mRNA-processing
factor SLU7
Length=585
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 99/215 (46%), Positives = 138/215 (64%), Gaps = 4/215 (1%)
Query 5 DKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQI 64
D P + D + RI R+LRIRED AKYL NLD NSA+YDPK+R+MRE+P+ + +
Sbjct 245 DDIDMPGQNFDSKRRITVRNLRIREDIAKYLRNLDPNSAYYDPKTRAMRENPYANAGKNP 304
Query 65 -KPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQI 123
+ ++ GDN + G+ + Q QLFAWEAY G VH A PT+LE LYK + KK
Sbjct 305 DEVSYAGDNFVRYTGDTISMAQTQLFAWEAYDKGSEVHLQADPTKLELLYKSFKVKKEDF 364
Query 124 ETEKQKNIFDTYGGKEHLVA-DTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVL 182
+ +++++I + YGG+EHL A +L AQTE YVEYS+ G +++G++R + SKYEEDV
Sbjct 365 KEQQKESILEKYGGQEHLDAPPAELLLAQTEDYVEYSRHGTVIKGQERAVACSKYEEDVK 424
Query 183 QGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
+HT +WGSY+ +WGY CC + CTG
Sbjct 425 INNHTHIWGSYW--KEGRWGYKCCHSFFKYSYCTG 457
> hsa:10569 SLU7, 9G8, MGC9280, hSlu7; SLU7 splicing factor homolog
(S. cerevisiae); K12819 pre-mRNA-processing factor SLU7
Length=586
Score = 197 bits (500), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 99/215 (46%), Positives = 138/215 (64%), Gaps = 4/215 (1%)
Query 5 DKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQI 64
D P + D + RI R+LRIRED AKYL NLD NSA+YDPK+R+MRE+P+ + +
Sbjct 245 DDIDMPGQNFDSKRRITVRNLRIREDIAKYLRNLDPNSAYYDPKTRAMRENPYANAGKNP 304
Query 65 -KPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQI 123
+ ++ GDN + G+ + Q QLFAWEAY G VH A PT+LE LYK + KK
Sbjct 305 DEVSYAGDNFVRYTGDTISMAQTQLFAWEAYDKGSEVHLQADPTKLELLYKSFKVKKEDF 364
Query 124 ETEKQKNIFDTYGGKEHLVA-DTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVL 182
+ +++++I + YGG+EHL A +L AQTE YVEYS+ G +++G++R + SKYEEDV
Sbjct 365 KEQQKESILEKYGGQEHLDAPPAELLLAQTEDYVEYSRHGTVIKGQERAVACSKYEEDVK 424
Query 183 QGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
+HT +WGSY+ +WGY CC + CTG
Sbjct 425 IHNHTHIWGSYW--KEGRWGYKCCHSFFKYSYCTG 457
> cel:K07C5.6 hypothetical protein; K12819 pre-mRNA-processing
factor SLU7
Length=647
Score = 191 bits (486), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 95/208 (45%), Positives = 144/208 (69%), Gaps = 7/208 (3%)
Query 15 DERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLS-QQIKPT-FRGDN 72
D R+RI R+LRIREDTAKYL NL NS +YDPKSRSMRE+PF ++ ++++ F GDN
Sbjct 230 DSRTRITVRNLRIREDTAKYLYNLAENSPYYDPKSRSMRENPFAGVAGKELEAARFSGDN 289
Query 73 ALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIF 132
+ +GEV A + Q+FAW+A + G H A+PT+LE L KE++K+K+ ++ E QK +
Sbjct 290 FVRYSGEVTAANEAQVFAWQATRGGVYAHSIAEPTKLEALKKEYEKEKSTLKNETQKELL 349
Query 133 DTYGGKEHL--VADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQGSHTSVW 190
D YGG EH+ AD +L AQTE+Y+EY++ G++++G+++ S+++ED+ +HTSV+
Sbjct 350 DKYGGGEHMERPAD-ELLLAQTESYIEYNRKGKVIKGKEKVAISSRFKEDIYPQNHTSVF 408
Query 191 GSYYDVATSKWGYACCRLTSFAAECTGR 218
GS++ +WGY CC + CTG+
Sbjct 409 GSFW--REGRWGYKCCHQFVKNSYCTGK 434
> ath:AT1G65660 SMP1; SMP1 (SWELLMAP 1); nucleic acid binding
/ single-stranded RNA binding; K12819 pre-mRNA-processing factor
SLU7
Length=535
Score = 179 bits (453), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 92/198 (46%), Positives = 124/198 (62%), Gaps = 6/198 (3%)
Query 22 TRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGDNALLQAGEVK 81
R+LRIREDTAKYLLNLD+NSA YDPK+RSMREDP K + GDN +G+
Sbjct 235 VRNLRIREDTAKYLLNLDVNSAHYDPKTRSMREDPLPDADPNDK-FYLGDNQYRNSGQAL 293
Query 82 ATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIFDTYG--GKE 139
+QL + +WEA+ GQ++H A P+Q E LYK Q K +++++ + I D YG E
Sbjct 294 EFKQLNIHSWEAFDKGQDMHMQAAPSQAELLYKSFQVAKEKLKSQTKDTIMDKYGNAATE 353
Query 140 HLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQGSHTSVWGSYYDVATS 199
+ +L Q+E VEY + GRI++G++ L KSKYEEDV +HTSVWGSY+
Sbjct 354 DEIP-MELLLGQSERQVEYDRAGRIIKGQEVILPKSKYEEDVHANNHTSVWGSYW--KDH 410
Query 200 KWGYACCRLTSFAAECTG 217
+WGY CC+ + CTG
Sbjct 411 QWGYKCCQQIIRNSYCTG 428
> bbo:BBOV_II001930 18.m06149; mRNA processing-related protein;
K12819 pre-mRNA-processing factor SLU7
Length=388
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 88/205 (42%), Positives = 125/205 (60%), Gaps = 16/205 (7%)
Query 14 TDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGDNA 73
+D+RSR +LRIREDTAKYL+NL+++SAFYDPKSR +R+DP +S TFRGDNA
Sbjct 196 SDDRSRNTMPNLRIREDTAKYLINLNIDSAFYDPKSRCLRDDPLLGMSNSDHHTFRGDNA 255
Query 74 LLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIFD 133
L +GE +++ FAWEA + G NV F AQPT+LEF++K+ K+ + + K++++ D
Sbjct 256 LFTSGEASRPGEIEQFAWEAQQKGSNVSFMAQPTELEFMFKDSSLKREEAKASKRQSLLD 315
Query 134 TYGGKEHLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLT-KSKYEEDVLQGSHTSVWGS 192
+GG AY+ D ++ ++ ++ E DV+ HTSVWGS
Sbjct 316 RFGGS---------------AYIRKDGDEDLIDTNLPVVSHTTEKETDVMLLGHTSVWGS 360
Query 193 YYDVATSKWGYACCRLTSFAAECTG 217
YYD +T WGY CC TS + CT
Sbjct 361 YYDRSTGLWGYKCCLSTSNTSRCTA 385
> pfa:PFF0500c step II splicing factor, putative; K12819 pre-mRNA-processing
factor SLU7
Length=444
Score = 172 bits (436), Expect = 8e-43, Method: Compositional matrix adjust.
Identities = 101/214 (47%), Positives = 137/214 (64%), Gaps = 13/214 (6%)
Query 13 CT---DERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPT-- 67
CT +E++R R+LRIREDTAKYL NL LNSAFYDPKSRSMREDPF ++ + +
Sbjct 230 CTINKNEKNRNIARNLRIREDTAKYLYNLSLNSAFYDPKSRSMREDPFANIRKHLNDDDN 289
Query 68 -FRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETE 126
++G+N + +++L++FAWE+YK G+NVHFNAQPTQLE +YKE+ +KK +I +
Sbjct 290 YYKGENYYNNTDDAIESKKLEVFAWESYKRGENVHFNAQPTQLELMYKEYLEKKKKIIKK 349
Query 127 KQKNIFDTY---GGKEHLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTK--SKYEEDV 181
KQ++I TY +E +L Q+E Y EY +I + K SKYEED+
Sbjct 350 KQEDILKTYKCQNNEEQQPNQEELL--QSEVYTEYKPIEQIHNNNMKKNIKVPSKYEEDI 407
Query 182 LQGSHTSVWGSYYDVATSKWGYACCRLTSFAAEC 215
H+SV+GSYYD T KWGY CC T+ +C
Sbjct 408 YLFDHSSVFGSYYDKHTKKWGYKCCSSTNKYDKC 441
> ath:AT4G37120 SMP2; SMP2; single-stranded RNA binding
Length=536
Score = 167 bits (422), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 86/198 (43%), Positives = 123/198 (62%), Gaps = 6/198 (3%)
Query 22 TRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGDNALLQAGEVK 81
R+LRIREDTAKYLLNLD+NSA YDPK+RSMREDP K + GDN +G+
Sbjct 235 VRNLRIREDTAKYLLNLDVNSAHYDPKTRSMREDPLPDADPNEK-FYLGDNQYRNSGQAL 293
Query 82 ATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNIFDTYG--GKE 139
+Q+ + + EA+ G ++H A P+Q E LYK + K +++T+ + I + YG E
Sbjct 294 EFKQINIHSCEAFDKGHDMHMQAAPSQAELLYKNFKVAKEKLKTQTKDTIMEKYGNAATE 353
Query 140 HLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQGSHTSVWGSYYDVATS 199
+ +L Q+E +EY + GRI++G++ + KSKYEEDV +HTSVWGS++
Sbjct 354 GEIP-MELLLGQSERQIEYDRAGRIMKGQEVIIPKSKYEEDVHANNHTSVWGSWW--KDH 410
Query 200 KWGYACCRLTSFAAECTG 217
+WGY CC+ T + CTG
Sbjct 411 QWGYKCCQQTIRNSYCTG 428
> cpv:cgd7_1750 step II splicing factor SLU7 ; K12819 pre-mRNA-processing
factor SLU7
Length=405
Score = 161 bits (408), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 90/217 (41%), Positives = 121/217 (55%), Gaps = 6/217 (2%)
Query 1 LKEFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHL 60
L +FD+T+ + +++R N R+LRIREDTAKYL NLDLNSAFYDPKSRSMREDPF H
Sbjct 194 LGDFDETT--FGASSDKTRTNVRNLRIREDTAKYLRNLDLNSAFYDPKSRSMREDPF-HK 250
Query 61 SQQIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKK 120
S I T+RGDNA+ +GEV ++ FA+ +K G+++H A PT+ E LYK K
Sbjct 251 SSNIGNTYRGDNAIRNSGEVSKILLMEAFAYNKHKQGESIHLQALPTRSEMLYKSSISNK 310
Query 121 AQIETEKQKNIFDTYGGKEHLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEED 180
+K + I Y L + + S + + S Y ED
Sbjct 311 DNDNLKKLEEISRKYERNNQPSISKLELTERPTTVGDLSS---MNTKTTKKFVSSIYVED 367
Query 181 VLQGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
+HT VWGSYYD+ KWG+ CC+ T ++CT
Sbjct 368 EYISNHTQVWGSYYDLEAKKWGFRCCKQTCRFSKCTN 404
> tpv:TP04_0325 step II splicing factor; K12819 pre-mRNA-processing
factor SLU7
Length=387
Score = 152 bits (385), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 80/206 (38%), Positives = 122/206 (59%), Gaps = 16/206 (7%)
Query 12 ECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFRGD 71
+C D+++R TR+LRIREDTAKYL+NLD+NSAFYDPKSRSMREDP + F+GD
Sbjct 196 DCKDDKTRTTTRNLRIREDTAKYLINLDVNSAFYDPKSRSMREDPL----LGVNTCFKGD 251
Query 72 NALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQKNI 131
N + E ++L++FAWE+ G +V F A PT+LE L+ E +++K + E ++ +
Sbjct 252 NYYFNSEETYKPKELEVFAWESKSKGVDVDFIANPTKLEKLFNETKERKEKETLESKQKL 311
Query 132 FDTYGGKEHLVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVLQG-SHTSVW 190
+ + E++ Y + + +K+ I T +S+YE D + H+ +W
Sbjct 312 IERFKASEYVNN-----YEELKPLSTVTKEDII------TFKESEYEFDEAKMLGHSQIW 360
Query 191 GSYYDVATSKWGYACCRLTSFAAECT 216
GS+YDV WGY CC+ T+ + C
Sbjct 361 GSFYDVEKGLWGYKCCKSTNRSQRCN 386
> ath:AT3G45950 splicing factor-related
Length=385
Score = 109 bits (273), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 57/134 (42%), Positives = 80/134 (59%), Gaps = 1/134 (0%)
Query 3 EFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQ 62
+F K V TD S+ R+LRIRED AKYLLNLD+NSA+YDPKSRSMREDP +
Sbjct 206 DFAKVKKRVRTTDGGSKGTVRNLRIREDPAKYLLNLDVNSAYYDPKSRSMREDPLPYTDP 265
Query 63 QIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQ 122
K R DN +G+ +Q +++ EA+ GQ++H A P+Q E YK + K +
Sbjct 266 NEKFCLR-DNQYRNSGQAIEFKQQNMYSCEAFDKGQDIHMQAAPSQAELCYKRVKIAKEK 324
Query 123 IETEKQKNIFDTYG 136
+ ++++ I YG
Sbjct 325 LNSQRKDAIIAKYG 338
> tpv:TP01_1089 hypothetical protein
Length=218
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 47/93 (50%), Gaps = 16/93 (17%)
Query 139 EHLVADTRVLYAQTEAYVEYSKDGRILRG----------------RQRTLTKSKYEEDVL 182
E ++ + R Y +V ++ G IL G ++ + +S+Y EDVL
Sbjct 77 EMILEEKRNKYKVDRNFVSENQSGLILTGPATSSTSPTPKSTNKEEKKLVIRSQYNEDVL 136
Query 183 QGSHTSVWGSYYDVATSKWGYACCRLTSFAAEC 215
+ HT+V+GSYYD WGY CC+LT A+C
Sbjct 137 KNGHTTVFGSYYDRDKKSWGYKCCKLTEKEAKC 169
> cpv:cgd7_1180 hypothetical protein
Length=227
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 177 YEEDVLQGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
YEEDV G+H+SVWGSYYD KWG+ CC+ ++ C+
Sbjct 111 YEEDVFLGNHSSVWGSYYDPNVKKWGFKCCKNLKRSSVCSA 151
> tgo:TGME49_063310 hypothetical protein
Length=288
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 177 YEEDVLQGSHTSVWGSYYDVATSKWGYACCRL 208
YEEDV GSHT VWGS++D AT++WG+ CC +
Sbjct 120 YEEDVYVGSHTHVWGSFFDKATNRWGFKCCGI 151
> pfa:MAL13P1.242 step II splicing factor, putative
Length=204
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 169 QRTLTKSKYEEDVLQGSHTSVWGSYYDVATSKWGYACCRLTSFAAECTG 217
+ L +SKY+ED+ +H S++GSYYD +KWG+ CC ++CT
Sbjct 113 NKILIRSKYDEDIFLKNHRSIYGSYYDKEKNKWGFKCCTNIDKNSKCTN 161
> hsa:80208 SPG11, DKFZp762B1512, FLJ21439, KIAA1840; spastic
paraplegia 11 (autosomal recessive)
Length=2330
Score = 33.5 bits (75), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 37/142 (26%), Positives = 58/142 (40%), Gaps = 22/142 (15%)
Query 1 LKEFDKTSAPVECTDERSRINTRDLRIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHL 60
L + + S P+ + + I R L DT + L N ++P S+S D F HL
Sbjct 520 LNGWGRCSIPIHALE--AGIENRQL----DTVNFFLKSKEN--LFNPSSKSSVSDQFDHL 571
Query 61 SQQIKPTFRGDNALLQAGEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKK 120
S + R L+ A ++ + E+Y Q+ HF+ Q L + +Q K+
Sbjct 572 SSHL--YLRNVEELIPALDLLCSA-----IRESYSEPQSKHFSEQLLNLTLSFLNNQIKE 624
Query 121 AQIETE-------KQKNIFDTY 135
I TE K NI +Y
Sbjct 625 LFIHTEELDEHLQKGVNILTSY 646
> mmu:237339 L3mbtl3, AI481284, MBT-1, MGC90546; l(3)mbt-like
3 (Drosophila)
Length=883
Score = 33.1 bits (74), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 31/61 (50%), Gaps = 6/61 (9%)
Query 141 LVADTRVLYAQTEAYVEYSKDGRILRGRQRTLTKSKYEEDVL------QGSHTSVWGSYY 194
L AD++ + + +E +DGRILRG QR K + + VL +G T W SY
Sbjct 179 LKADSKDDGEERDDEMENKQDGRILRGSQRARRKRRGDSAVLKQGLPPKGKKTWCWASYL 238
Query 195 D 195
+
Sbjct 239 E 239
> sce:YDR088C SLU7, SLT17; RNA splicing factor, required for ATP-independent
portion of 2nd catalytic step of spliceosomal
RNA splicing; interacts with Prp18p; contains zinc knuckle
domain; K12819 pre-mRNA-processing factor SLU7
Length=382
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 55/112 (49%), Gaps = 14/112 (12%)
Query 26 RIREDTAKYLLNLDLNSAFYDPKSRSMREDPFKHLSQQIKPTFR----GD----NALLQA 77
R+RED A YL +++ + YDPKSR + + + ++ K FR G+ N L Q
Sbjct 247 RLREDKAAYLNDINSTESNYDPKSRLYKTETLGAVDEKSK-MFRRHLTGEGLKLNELNQF 305
Query 78 GEVKATQQLQLFAWEAYKHGQNVHFNAQPTQLEFLYKEHQKKKAQIETEKQK 129
A + E + Q+V A PT+ E+L +KK+ Q ET++ K
Sbjct 306 ARSHAKEMGIRDEIEDKEKVQHV-LVANPTKYEYL----KKKREQEETKQPK 352
Lambda K H
0.316 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7395169300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40