bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
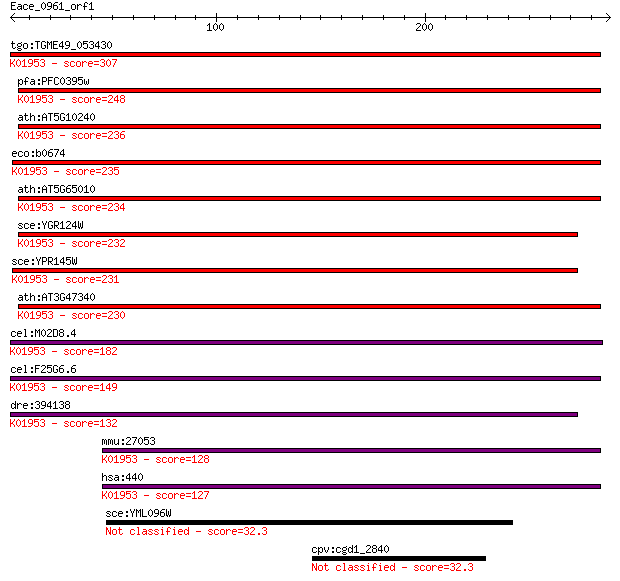
Query= Eace_0961_orf1
Length=288
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_053430 asparagine synthase, putative (EC:6.3.5.4); ... 307 2e-83
pfa:PFC0395w asparagine synthetase, putative (EC:6.3.5.4); K01... 248 2e-65
ath:AT5G10240 ASN3; ASN3 (ASPARAGINE SYNTHETASE 3); asparagine... 236 1e-61
eco:b0674 asnB, ECK0662, JW0660; asparagine synthetase B (EC:6... 235 1e-61
ath:AT5G65010 ASN2; ASN2 (ASPARAGINE SYNTHETASE 2); asparagine... 234 3e-61
sce:YGR124W ASN2; Asn2p (EC:6.3.5.4); K01953 asparagine syntha... 232 1e-60
sce:YPR145W ASN1; Asn1p (EC:6.3.5.4); K01953 asparagine syntha... 231 3e-60
ath:AT3G47340 ASN1; ASN1 (GLUTAMINE-DEPENDENT ASPARAGINE SYNTH... 230 5e-60
cel:M02D8.4 hypothetical protein; K01953 asparagine synthase (... 182 1e-45
cel:F25G6.6 nrs-2; asparagiNyl tRNA Synthetase family member (... 149 9e-36
dre:394138 asns, MGC56605, zgc:56605, zgc:76901; asparagine sy... 132 1e-30
mmu:27053 Asns; asparagine synthetase (EC:6.3.5.4); K01953 asp... 128 2e-29
hsa:440 ASNS, TS11; asparagine synthetase (glutamine-hydrolyzi... 127 6e-29
sce:YML096W Putative protein of unknown function with similari... 32.3 2.1
cpv:cgd1_2840 contains a UVDDB domain that is present in CPSF_... 32.3 2.1
> tgo:TGME49_053430 asparagine synthase, putative (EC:6.3.5.4);
K01953 asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=588
Score = 307 bits (787), Expect = 2e-83, Method: Compositional matrix adjust.
Identities = 141/283 (49%), Positives = 200/283 (70%), Gaps = 0/283 (0%)
Query 1 RVREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMM 60
+V FPPGH + ++ G +R+Y P W + P+P+ CDL +LRE LE AV KR+M
Sbjct 197 QVTVFPPGHFYLSTMNEGKGGFQRYYNPSWWGVDKPLPTYRCDLGQLREALEAAVRKRLM 256
Query 61 CDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSR 120
CDVPFG++L GG+DS+IIA+I + + + E+ H WT +IH+F+IGLK D + ++
Sbjct 257 CDVPFGLMLSGGVDSSIIAAIAAKEFQKMSATEKGSHIWTNQIHAFSIGLKDGPDRKFAK 316
Query 121 MVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMV 180
MVA +G HHEFTFE +E +DAL D+IYMIE+YD+ TVRA++ Y L R+IKS+GVK+V
Sbjct 317 MVADMLGTVHHEFTFELEEGLDALDDVIYMIETYDITTVRASVPMYLLCRMIKSLGVKVV 376
Query 181 ITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYL 240
+TGEGA ++FG + EA H+ + +++ +HF D LRANK SM WGVEARVP+L
Sbjct 377 LTGEGADEVFGGYLYFHSAPNREAFHRELQNKLNLVHFYDCLRANKTSMAWGVEARVPFL 436
Query 241 DVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEELPQFIIDR 283
D F+D+AMSIDP++K C+ GR+ KQ+LR+A+K LP ++ R
Sbjct 437 DRGFLDFAMSIDPEDKMCTNGRMEKQILRDAFKGYLPDEVLYR 479
> pfa:PFC0395w asparagine synthetase, putative (EC:6.3.5.4); K01953
asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=610
Score = 248 bits (633), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 128/307 (41%), Positives = 190/307 (61%), Gaps = 30/307 (9%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH Y+K++K G R+Y P W IP+ D + +R LEKAV KR+M DVP
Sbjct 182 FPPGH--YYKNNKNKGEFVRYYNPNWWSLNNSIPNNKVDFNEIRIHLEKAVIKRLMGDVP 239
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKE----QNEH----------------------- 97
FG+LL GG+DS+IIA+I+ + N NK QN +
Sbjct 240 FGILLSGGLDSSIIAAILAKHLNILDNKNGKSIQNGNDKKSNNNGNESNNNNNNNNFNNN 299
Query 98 -FWTPKIHSFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDV 156
K+ SF+IGLKGS DL+ ++ VA+++G H EF F +E ID+L D+IY IE+YD+
Sbjct 300 NSGPQKLRSFSIGLKGSPDLKAAKEVAEYLGIEHTEFYFTVEEGIDSLHDVIYHIETYDI 359
Query 157 LTVRAALLNYFLYRLIKSIGVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDM 216
T+RA+ Y L RLIKS VKMV++GEG+ ++FG + + E H+ + +++ D+
Sbjct 360 TTIRASTPMYILSRLIKSSCVKMVLSGEGSDEIFGGYLYFHKAPNKEEFHRELQRKIHDL 419
Query 217 HFTDSLRANKASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEEL 276
H+ D LRANK++M +G+EARVP+LD+ F++ M+IDP++K CS ++ K +LR A++ L
Sbjct 420 HYYDVLRANKSTMAFGIEARVPFLDIQFLNVVMNIDPQDKMCSNNKIEKYILRKAFEGYL 479
Query 277 PQFIIDR 283
P+ I+ R
Sbjct 480 PEHILYR 486
> ath:AT5G10240 ASN3; ASN3 (ASPARAGINE SYNTHETASE 3); asparagine
synthase (glutamine-hydrolyzing) (EC:6.3.5.4); K01953 asparagine
synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=577
Score = 236 bits (601), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 126/285 (44%), Positives = 179/285 (62%), Gaps = 16/285 (5%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH + SSK G + R+Y P W +PS D +R EKAV KR+M DVP
Sbjct 173 FPPGHIY---SSKQGG-LRRWYNPPWFS--EVVPSTPYDPLVVRNTFEKAVIKRLMTDVP 226
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVAK 124
FGVLL GG+DS+++AS+ R K + W K+H+F IGLKGS DL+ R VA
Sbjct 227 FGVLLSGGLDSSLVASVALR----HLEKSEAACQWGSKLHTFCIGLKGSPDLKAGREVAD 282
Query 125 HVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVITGE 184
++G HHE F Q+ IDA+ ++IY +E+YDV T+RA+ + + R IKS+GVKMV++GE
Sbjct 283 YLGTRHHELHFTVQDGIDAIEEVIYHVETYDVTTIRASTPMFLMSRKIKSLGVKMVLSGE 342
Query 185 GAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVSF 244
G+ ++FG + + + H+ +++ +H D LRANK++ WGVEARVP+LD F
Sbjct 343 GSDEIFGGYLYFHKAPNKKEFHEETCRKIKALHQYDCLRANKSTSAWGVEARVPFLDKEF 402
Query 245 VDYAMSIDPKEK--RCSGGRLPKQLLRNAYKEE----LPQFIIDR 283
++ AMSIDP+ K R GR+ K +LRNA+ +E LP+ I+ R
Sbjct 403 INVAMSIDPEWKMIRPDLGRIEKWVLRNAFDDEKNPYLPKHILYR 447
> eco:b0674 asnB, ECK0662, JW0660; asparagine synthetase B (EC:6.3.5.4);
K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=554
Score = 235 bits (600), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 113/282 (40%), Positives = 174/282 (61%), Gaps = 5/282 (1%)
Query 2 VREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMC 61
++EFP G + + G + +Y W +A + D + LR+ LE +V +M
Sbjct 172 IKEFPAGSYLWSQD----GEIRSYYHRDWFDYDA-VKDNVTDKNELRQALEDSVKSHLMS 226
Query 62 DVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRM 121
DVP+GVLL GG+DS+II++I + R ++ W P++HSFA+GL GS DL+ ++
Sbjct 227 DVPYGVLLSGGLDSSIISAITKKYAARRVEDQERSEAWWPQLHSFAVGLPGSPDLKAAQE 286
Query 122 VAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVI 181
VA H+G HHE F QE +DA+ D+IY IE+YDV T+RA+ Y + R IK++G+KMV+
Sbjct 287 VANHLGTVHHEIHFTVQEGLDAIRDVIYHIETYDVTTIRASTPMYLMSRKIKAMGIKMVL 346
Query 182 TGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLD 241
+GEG+ ++FG + + + LH+ V+++ +H D RANKA WGVEARVP+LD
Sbjct 347 SGEGSDEVFGGYLYFHKAPNAKELHEETVRKLLALHMYDCARANKAMSAWGVEARVPFLD 406
Query 242 VSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEELPQFIIDR 283
F+D AM I+P++K C G++ K +LR ++ LP + R
Sbjct 407 KKFLDVAMRINPQDKMCGNGKMEKHILRECFEAYLPASVAWR 448
> ath:AT5G65010 ASN2; ASN2 (ASPARAGINE SYNTHETASE 2); asparagine
synthase (glutamine-hydrolyzing) (EC:6.3.5.4); K01953 asparagine
synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=579
Score = 234 bits (597), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 124/285 (43%), Positives = 180/285 (63%), Gaps = 16/285 (5%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH + SSK G + R+Y P W + +PS D LR EKAV KR+M DVP
Sbjct 174 FPPGHIY---SSKQGG-LRRWYNPPWYNEQ--VPSTPYDPLVLRNAFEKAVIKRLMTDVP 227
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVAK 124
FGVLL GG+DS+++A++ R K + W ++H+F IGL+GS DL+ R VA
Sbjct 228 FGVLLSGGLDSSLVAAVALR----HLEKSEAARQWGSQLHTFCIGLQGSPDLKAGREVAD 283
Query 125 HVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVITGE 184
++G HHEF F Q+ IDA+ ++IY IE+YDV T+RA+ + + R IKS+GVKMV++GE
Sbjct 284 YLGTRHHEFQFTVQDGIDAIEEVIYHIETYDVTTIRASTPMFLMSRKIKSLGVKMVLSGE 343
Query 185 GAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVSF 244
G+ ++ G + + + H+ +++ +H D LRANK++ WGVEARVP+LD F
Sbjct 344 GSDEILGGYLYFHKAPNKKEFHEETCRKIKALHQFDCLRANKSTSAWGVEARVPFLDKEF 403
Query 245 VDYAMSIDPKEK--RCSGGRLPKQLLRNAYKEE----LPQFIIDR 283
++ AMSIDP+ K + GR+ K +LRNA+ +E LP+ I+ R
Sbjct 404 LNVAMSIDPEWKLIKPDLGRIEKWVLRNAFDDEERPYLPKHILYR 448
> sce:YGR124W ASN2; Asn2p (EC:6.3.5.4); K01953 asparagine synthase
(glutamine-hydrolysing) [EC:6.3.5.4]
Length=572
Score = 232 bits (591), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 128/291 (43%), Positives = 179/291 (61%), Gaps = 31/291 (10%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH + ++ K + R++ P W+ E IPS D +R LEKAV KR+M +VP
Sbjct 175 FPPGHVYDSETDK----ITRYFTPDWLD-EKRIPSTPVDYHAIRHSLEKAVRKRLMAEVP 229
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKEQNE----------------HFWTP---KIHS 105
+GVLL GG+DS++IA+I R E+ N + NE H T ++HS
Sbjct 230 YGVLLSGGLDSSLIAAIAAR-ETEKANADANEDNNVDEKQLAGIDDQGHLHTSGWSRLHS 288
Query 106 FAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLN 165
FAIGL + DL+ +R VAK +G HHE TF QE +DAL D+IY +E+YDV T+RA+
Sbjct 289 FAIGLPNAPDLQAARKVAKFIGSIHHEHTFTLQEGLDALDDVIYHLETYDVTTIRASTPM 348
Query 166 YFLYRLIKSIGVKMVITGEGAADLFGCAGDMQEEQDPEA--LHKAIVKRMSDMHFTDSLR 223
+ L R IK+ GVKMV++GEG+ ++FG G + Q P A H V+R+ ++H D LR
Sbjct 349 FLLSRKIKAQGVKMVLSGEGSDEIFG--GYLYFAQAPSAAEFHTESVQRVKNLHLADCLR 406
Query 224 ANKASMCWGVEARVPYLDVSFVDYAMSIDPKEK--RCSGGRLPKQLLRNAY 272
ANK++M WG+EARVP+LD F+ M+IDP EK + GR+ K +LR A+
Sbjct 407 ANKSTMAWGLEARVPFLDKDFLQLCMNIDPNEKMIKPKEGRIEKYILRKAF 457
> sce:YPR145W ASN1; Asn1p (EC:6.3.5.4); K01953 asparagine synthase
(glutamine-hydrolysing) [EC:6.3.5.4]
Length=572
Score = 231 bits (588), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 127/295 (43%), Positives = 177/295 (60%), Gaps = 32/295 (10%)
Query 2 VREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMC 61
+ FPPGH + K+ K + R++ P W+ E IPS D +R LEKAV KR+M
Sbjct 172 ITAFPPGHVYDSKTDK----ITRYFTPDWLD-EKRIPSTPIDYMAIRHSLEKAVRKRLMA 226
Query 62 DVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHF--------------------WTP 101
+VP+GVLL GG+DS++IASI R + N + + WT
Sbjct 227 EVPYGVLLSGGLDSSLIASIAARETAKATNDVEPSTYDSKARHLAGIDDDGKLHTAGWT- 285
Query 102 KIHSFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRA 161
+HSFAIGL + DL+ +R VAK +G HHE TF QE +DAL D+IY +E+YDV T+RA
Sbjct 286 SLHSFAIGLPNAPDLQAARKVAKFIGSIHHEHTFTLQEGLDALDDVIYHLETYDVTTIRA 345
Query 162 ALLNYFLYRLIKSIGVKMVITGEGAADLFGCAGDMQEEQDPEA--LHKAIVKRMSDMHFT 219
+ + L R IK+ GVKMV++GEG+ ++FG G + Q P A H V+R+ ++H
Sbjct 346 STPMFLLSRKIKAQGVKMVLSGEGSDEIFG--GYLYFAQAPSAAEFHTESVQRVKNLHLA 403
Query 220 DSLRANKASMCWGVEARVPYLDVSFVDYAMSIDPKEK--RCSGGRLPKQLLRNAY 272
D LRANK++M WG+EARVP+LD F+ M+IDP EK + GR+ K +LR A+
Sbjct 404 DCLRANKSTMAWGLEARVPFLDREFLQLCMNIDPNEKMIKPKEGRIEKYILRKAF 458
> ath:AT3G47340 ASN1; ASN1 (GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE
1); asparagine synthase (glutamine-hydrolyzing) (EC:6.3.5.4);
K01953 asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=584
Score = 230 bits (586), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 122/286 (42%), Positives = 182/286 (63%), Gaps = 18/286 (6%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH F SSK G +++Y P W +PS + +R E AV KR+M DVP
Sbjct 174 FPPGH---FYSSKLGGF-KQWYNPPWFNES--VPSTPYEPLAIRRAFENAVIKRLMTDVP 227
Query 65 FGVLLYGGIDSAIIASIVCR-LYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVA 123
FGVLL GG+DS+++ASI R L + K+ W P++HSF +GL+GS DL+ + VA
Sbjct 228 FGVLLSGGLDSSLVASITARHLAGTKAAKQ-----WGPQLHSFCVGLEGSPDLKAGKEVA 282
Query 124 KHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVITG 183
+++G HHEF F Q+ IDA+ D+IY +E+YDV T+RA+ + + R IKS+GVKMV++G
Sbjct 283 EYLGTVHHEFHFSVQDGIDAIEDVIYHVETYDVTTIRASTPMFLMSRKIKSLGVKMVLSG 342
Query 184 EGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVS 243
EGA ++FG + + + H+ +++ +H D LRANK++ +G+EARVP+LD
Sbjct 343 EGADEIFGGYLYFHKAPNKKEFHQETCRKIKALHKYDCLRANKSTSAFGLEARVPFLDKD 402
Query 244 FVDYAMSIDPKEK--RCSGGRLPKQLLRNAYKEE----LPQFIIDR 283
F++ AMS+DP+ K + GR+ K +LR A+ +E LP+ I+ R
Sbjct 403 FINTAMSLDPESKMIKPEEGRIEKWVLRRAFDDEERPYLPKHILYR 448
> cel:M02D8.4 hypothetical protein; K01953 asparagine synthase
(glutamine-hydrolysing) [EC:6.3.5.4]
Length=567
Score = 182 bits (462), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 106/289 (36%), Positives = 161/289 (55%), Gaps = 22/289 (7%)
Query 1 RVREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMM 60
++ FPPGH + K+ R++ P+W I DL L++ + +V KR+M
Sbjct 175 KIESFPPGHYYTPKTG-----FVRYFNPEWFDFRKAI--HPLDLKLLQKTMIASVHKRLM 227
Query 61 CDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGL-KGSRDLECS 119
D P GVLL GG+DS++++SI R R +HSF+IG+ S D+ +
Sbjct 228 SDAPIGVLLSGGLDSSLVSSIASREMKRR----------GMAVHSFSIGVDHNSPDVVAA 277
Query 120 RMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKM 179
R VAK +G THHEF F +E I L LI+ +ESYDV ++RA+ YFL I+ +G+K+
Sbjct 278 RKVAKFIGTTHHEFYFSIEEGIKNLRKLIWHLESYDVTSIRASTPMYFLSEEIRKLGIKV 337
Query 180 VITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPY 239
V++GEGA ++FG E K + R+ ++ +D LRA+K+SM VE RVP+
Sbjct 338 VLSGEGADEIFGGYLYFHNAPSDEDFQKETIDRVLHLYTSDCLRADKSSMAHSVEVRVPF 397
Query 240 LDVSFVDYAMSIDPKEKRCS----GGRLPKQLLRNAYKEELPQFIIDRI 284
LD +FV+ A+S+DP KR G K +LR+A+ + ++ D I
Sbjct 398 LDKAFVEAAVSLDPAFKRPQKLEDGRNCEKFVLRSAFNTDQYPYLPDEI 446
> cel:F25G6.6 nrs-2; asparagiNyl tRNA Synthetase family member
(nrs-2); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=551
Score = 149 bits (377), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 95/293 (32%), Positives = 156/293 (53%), Gaps = 33/293 (11%)
Query 1 RVREFPPGHSFYFKSSKASGIMERFYKPQWMQPEA--PIPSAA-------CDLSRLREEL 51
RV FPPG + F + +QP+ +P+ A C + +R+ L
Sbjct 167 RVEYFPPG---------CCATISLFAPTRPVQPQQYYSVPTIADRFLSIDCTKTLVRDVL 217
Query 52 EKAVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLK 111
K+V KR+M + FG +L GG+DS++IASI R F K +F++G +
Sbjct 218 VKSVEKRLMGNRNFGFMLSGGLDSSLIASIATR-------------FLKQKPIAFSVGFE 264
Query 112 GSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRL 171
S DLE ++ VA ++ H Q+ ID + ++++ +E++D L +R + +Y L +
Sbjct 265 DSPDLENAKKVADYLKIPHEVLVITPQQCIDIIPEVVFALETFDPLIIRCGIAHYLLCQH 324
Query 172 I-KSIGVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMC 230
I KS VK++++GEGA +LFG MQ + LHK I++RM +H D LR ++++ C
Sbjct 325 ISKSSDVKVLLSGEGADELFGSYAYMQRAPNALHLHKEILRRMHHLHQYDVLRCDRSTSC 384
Query 231 WGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEELPQFIIDR 283
G+E RVP+LD F+D + P K +L K +LR+A++ LP ++ R
Sbjct 385 HGLEIRVPFLDKRFIDLVSRLPPSYKLMP-MKLEKHVLRSAFEGWLPDEVLWR 436
> dre:394138 asns, MGC56605, zgc:56605, zgc:76901; asparagine
synthetase (EC:6.3.5.4); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=560
Score = 132 bits (333), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 93/287 (32%), Positives = 150/287 (52%), Gaps = 25/287 (8%)
Query 1 RVREFPPGHSFYFK---SSKASGI-MERFY----KPQWMQPEAPIPSAACDL------SR 46
++ FPPGH F + K + M+RF+ KP+ + D S
Sbjct 175 KITAFPPGHFEVFDLKLNGKVESVQMDRFHCCTDKPKHADFNK-LEGLGTDFELETVKSN 233
Query 47 LREELEKAVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSF 106
+R E AV KR+M G LL GG+DS+++A+++ +L E E P I +F
Sbjct 234 IRILFEDAVRKRLMAHRRIGCLLSGGLDSSLVAALLVKLAKE-------EKLPYP-IQTF 285
Query 107 AIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNY 166
+IG + S D+ +R VA ++G HH F +E I AL D+I +ESYD+ TVRA++ Y
Sbjct 286 SIGAEDSPDVAAARKVADYIGSEHHVVNFTPEEGISALEDVIVHLESYDITTVRASVGMY 345
Query 167 FLYRLIKSIGVKMVI-TGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRAN 225
+ + I+ +VI +GEG+ +L + P+A + V+ + +++ D LRA+
Sbjct 346 LVSKYIREKTDSVVIFSGEGSDELTQGYIYFHKAPSPKAGAEDSVRLLEELYLFDVLRAD 405
Query 226 KASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAY 272
+ + G+E RVP+LD F Y +S+ P+E R + K LLR+A+
Sbjct 406 RTTAAHGLELRVPFLDHRFTAYYLSL-PEEMRVPKNGVEKHLLRDAF 451
> mmu:27053 Asns; asparagine synthetase (EC:6.3.5.4); K01953 asparagine
synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=561
Score = 128 bits (322), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 76/242 (31%), Positives = 137/242 (56%), Gaps = 12/242 (4%)
Query 45 SRLREELEKAVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIH 104
+ LR + A+ KR+M D G LL GG+DS+++A+ + + + + Q ++ +
Sbjct 233 NNLRILFDNAIKKRLMTDRRIGCLLSGGLDSSLVAASLLK----QLKEAQVQY----PLQ 284
Query 105 SFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALL 164
+FAIG++ S DL +R VA ++G HHE F ++E I AL ++I+ +E+YD+ TVRA++
Sbjct 285 TFAIGMEDSPDLLAARKVANYIGSEHHEVLFNSEEGIQALDEVIFSLETYDITTVRASVG 344
Query 165 NYFLYRLI-KSIGVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLR 223
Y + + I K+ ++ +GEG+ +L + PE + + + +++ D LR
Sbjct 345 MYLISKYIRKNTDSVVIFSGEGSDELTQGYIYFHKAPSPEKAEEESERLLKELYLFDVLR 404
Query 224 ANKASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKE--ELPQFII 281
A++ + G+E RVP+LD F Y +S+ P + R + K LLR +++ LP+ I+
Sbjct 405 ADRTTAAHGLELRVPFLDHRFSSYYLSL-PPDMRIPKNGIEKHLLRETFEDCNLLPKEIL 463
Query 282 DR 283
R
Sbjct 464 WR 465
> hsa:440 ASNS, TS11; asparagine synthetase (glutamine-hydrolyzing)
(EC:6.3.5.4); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=540
Score = 127 bits (318), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 77/242 (31%), Positives = 136/242 (56%), Gaps = 12/242 (4%)
Query 45 SRLREELEKAVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIH 104
+ LR AV KR+M D G LL GG+DS+++A+ + + + + Q ++ +
Sbjct 212 NNLRILFNNAVKKRLMTDRRIGCLLSGGLDSSLVAATLLK----QLKEAQVQY----PLQ 263
Query 105 SFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALL 164
+FAIG++ S DL +R VA H+G H+E F ++E I AL ++I+ +E+YD+ TVRA++
Sbjct 264 TFAIGMEDSPDLLAARKVADHIGSEHYEVLFNSEEGIQALDEVIFSLETYDITTVRASVG 323
Query 165 NYFLYRLI-KSIGVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLR 223
Y + + I K+ ++ +GEG+ +L + PE + + + +++ D LR
Sbjct 324 MYLISKYIRKNTDSVVIFSGEGSDELTQGYIYFHKAPSPEKAEEESERLLRELYLFDVLR 383
Query 224 ANKASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEE--LPQFII 281
A++ + G+E RVP+LD F Y +S+ P E R + K LLR +++ +P+ I+
Sbjct 384 ADRTTAAHGLELRVPFLDHRFSSYYLSL-PPEMRIPKNGIEKHLLRETFEDSNLIPKEIL 442
Query 282 DR 283
R
Sbjct 443 WR 444
> sce:YML096W Putative protein of unknown function with similarity
to asparagine synthetases; green fluorescent protein (GFP)-fusion
protein localizes to the cytoplasm; YML096W is not
an essential gene and partially overlaps the verified gene
RAD10
Length=525
Score = 32.3 bits (72), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 49/228 (21%), Positives = 96/228 (42%), Gaps = 40/228 (17%)
Query 47 LREELEK---AVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNK-------EQNE 96
LR+ ++K ++ R + + P VL GGID ++I +++C + E K N
Sbjct 234 LRDSVKKRVESIHPRHIENSPIAVLFSGGIDCSVIVALICEVLQENDYKCGKPVIELLNV 293
Query 97 HFWTPKIHSFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSD---------- 146
F P+ F + D + S AK + + + E +D D
Sbjct 294 SFENPRTGLFP---SDTPDRKLSINSAKTLQNLYPNVDIKLVE-VDVPYDEYLKWKPFVI 349
Query 147 -LIYMIESYDVLTVRAALLNYFLYR---LIKSIGVK---------MVITGEGAADLFGCA 193
L+Y ++ L++ A +F R + S+ + ++ +G GA +L+G
Sbjct 350 NLMYPKQTEMDLSIAIAF--FFASRGRGFLTSLNGERTPYQRHGIVLFSGLGADELYGGY 407
Query 194 GDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLD 241
+ P L + + +++++++ + R +K GVE R P+LD
Sbjct 408 HKFA-NKPPHELVEELTRQINNIYDRNLNRDDKVIAHNGVEVRYPFLD 454
> cpv:cgd1_2840 contains a UVDDB domain that is present in CPSF_A
and damage specific DNA binding protein 1
Length=1495
Score = 32.3 bits (72), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 46/109 (42%), Gaps = 26/109 (23%)
Query 146 DLIYMIESYDVLTVRAALLN--------------YFLYR--LIKSIGVKMVITGE----- 184
D++ Y+++ +RA LLN LYR L KS +KM++ E
Sbjct 96 DVLIFTREYELILLRAHLLNSDSEKILCMSILDKVSLYRENLRKSQLIKMMVHSEKNRIV 155
Query 185 -----GAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKAS 228
G + GC ++E+D ++ R+S++ TD AN +
Sbjct 156 ILAYEGCLQVVGCEISSKDEEDKAVFTSPLIIRLSELSVTDICLANTTN 204
Lambda K H
0.323 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11294782764
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40