bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0962_orf6
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
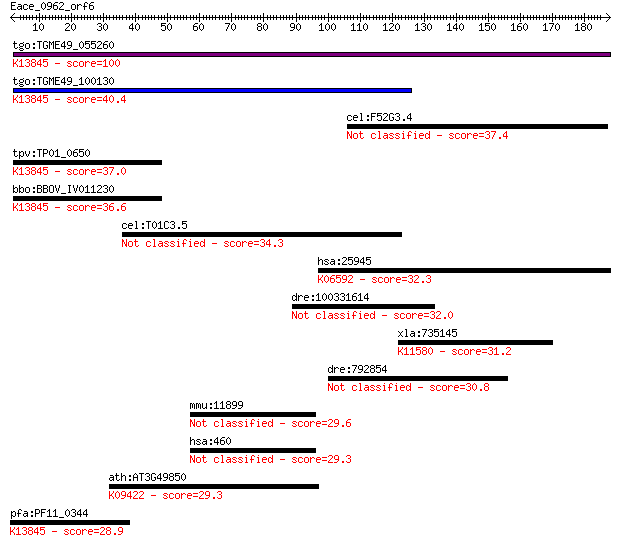
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 100 2e-21
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 40.4 0.003
cel:F52G3.4 hypothetical protein 37.4 0.028
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 37.0 0.037
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 36.6 0.051
cel:T01C3.5 hypothetical protein 34.3 0.28
hsa:25945 PVRL3, CD113, CDw113, DKFZp566B0846, FLJ90624, PPR3,... 32.3 1.1
dre:100331614 hypothetical protein LOC100331614 32.0 1.2
xla:735145 sgol1, MGC115705, sgo1; shugoshin-like 1; K11580 sh... 31.2 2.2
dre:792854 starch binding domain 1-like 30.8 3.1
mmu:11899 Astn1, Astn, GC14, mKIAA0289; astrotactin 1 29.6
hsa:460 ASTN1, ASTN, KIAA1747; astrotactin 1 29.3
ath:AT3G49850 TRB3; TRB3 (TELOMERE REPEAT BINDING FACTOR 3); D... 29.3 9.0
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 28.9 9.9
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 81/209 (38%), Positives = 104/209 (49%), Gaps = 30/209 (14%)
Query 2 GVNYGSYYPSSK---ECWLSDEVPNCLVPMDGAVAYTALGGLDEE--------------- 43
G NYG YY + +C LSD+VP+CLV AV+YTA G L EE
Sbjct 368 GRNYGFYYVDTTGEGKCALSDQVPDCLVSDSAAVSYTAAGSLSEETPNFIIPSNPSVTPP 427
Query 44 ----AAPCT-DSFPPTSTPCDPSTCTITLTSCVNGALVSKEVACPAEESNVCEGSRFSAV 98
A CT D FP + CD C TSCV G + S V C A+E N C GS + +
Sbjct 428 TPETALQCTADKFPDSFGACDVQACKRQKTSCVGGQIQSTSVDCTADEQNEC-GSNTALI 486
Query 99 MIGLAAAGGILLLLLAGGGFALYRSRRGPARKGEEATRSDYVQEEAAANRKKQRQSDLVQ 158
+L LL G FA +R KG +A ++ + RKK R SDL+Q
Sbjct 487 AGLAVGGVLLLALLGGGCYFA----KRLDRNKGVQAAHHEHEFQSDRGARKK-RPSDLMQ 541
Query 159 QAEPSFWEEAEADENEGAENTHVLVDQDY 187
+AEPSFW+EAE + + E THV+V+ DY
Sbjct 542 EAEPSFWDEAEENIEQDGE-THVMVEGDY 569
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 40/143 (27%), Positives = 60/143 (41%), Gaps = 33/143 (23%)
Query 2 GVNYGSYY--PSSKECWLSDEVPNCLVPMDGAVAYTALGGLDEEAAPCTDSFPPTSTPCD 59
G+N+ ++Y PS C + D VPNCL A+ +LG + P PP S+ +
Sbjct 365 GINWANFYKKPSGSYCEMYDGVPNCLTSAPEQYAFVSLG----DPNPDNAQLPPCSSATE 420
Query 60 ----PSTCTI---------TLTSCVNGALVSKEVACPAEESNVCEGSRFSAVMIGLAA-- 104
PS C+ + C +G V V+C C ++V I L A
Sbjct 421 GVVIPSHCSCPEGETSGSSSGVKCEDGKWVEGHVSC------TCSLDEDTSVNIWLIAGP 474
Query 105 --AGGILLLLLAGGGFALYRSRR 125
A G+LLL GG + ++R
Sbjct 475 CIAAGVLLL----GGLIYWMAQR 493
> cel:F52G3.4 hypothetical protein
Length=1009
Score = 37.4 bits (85), Expect = 0.028, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 39/83 (46%), Gaps = 2/83 (2%)
Query 106 GGILLLLLAG-GGFALYRSRRGPARKGEEATRSDYVQEEAAANRK-KQRQSDLVQQAEPS 163
GG +LL G GG+ +++ RR KGE T +V EE AA RK K+ D
Sbjct 440 GGYVLLFPDGVGGYNIHKFRRHDFSKGEPLTWEQHVLEEVAAIRKRKENPEDYYSNITEL 499
Query 164 FWEEAEADENEGAENTHVLVDQD 186
+ E E E+ V +++D
Sbjct 500 LRSQQEKKSAEQQEDQDVDMEED 522
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 37.0 bits (84), Expect = 0.037, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 2 GVNYGSYYPSSKECWLSDEVPNCLVPMDGAVAYTALGGLDEEAA---PC 47
G+N+ +Y K+C + D VP CL+ +G A T+L +E+ A PC
Sbjct 545 GINWATYSVEEKKCNILDVVPTCLIISNGYYALTSLSSPNEDDAINYPC 593
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 36.6 bits (83), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 27/49 (55%), Gaps = 3/49 (6%)
Query 2 GVNYGSYYPSSKECWLSDEVPNCLVPMDGAVAYTALGG---LDEEAAPC 47
G+N+ +Y S C L +E PNCL+ G++A TA+G D PC
Sbjct 398 GMNWATYDKDSGMCALINETPNCLILNAGSIALTAIGSPLEYDAVNYPC 446
> cel:T01C3.5 hypothetical protein
Length=243
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 40/88 (45%), Gaps = 1/88 (1%)
Query 36 ALGGLDEEAAPCTDSFPPTS-TPCDPSTCTITLTSCVNGALVSKEVACPAEESNVCEGSR 94
AL G DE+ D+FP + T S + L S + KE + + E S
Sbjct 121 ALNGKDEDVQFENDNFPSAAYTEVTSSRDLLILESKSRASHSRKECSAGFFQFKTEESSG 180
Query 95 FSAVMIGLAAAGGILLLLLAGGGFALYR 122
+A++IGL A I L + G G+ +Y+
Sbjct 181 MNAIVIGLIIACVIAFLAIIGQGYFMYK 208
> hsa:25945 PVRL3, CD113, CDw113, DKFZp566B0846, FLJ90624, PPR3,
PRR3, PVRR3, nectin-3; poliovirus receptor-related 3; K06592
poliovirus receptor-related protein 3
Length=549
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 43/94 (45%), Gaps = 13/94 (13%)
Query 97 AVMIGLAAAGGILLLLLAG-GGFALYRSRRGPARKGEEATRSDYVQEEAA--ANRKKQRQ 153
A +I G + ++L++ G YR RR R DY + ++ +K+ Q
Sbjct 403 ATIIASVVGGALFIVLVSVLAGIFCYRRRR--------TFRGDYFAKNYIPPSDMQKESQ 454
Query 154 SDLVQQAEPSFWEEAEADENEGAENTHVLVDQDY 187
D++QQ E + ++ EN+ N L+ +DY
Sbjct 455 IDVLQQDELDSYPDSVKKENKNPVNN--LIRKDY 486
> dre:100331614 hypothetical protein LOC100331614
Length=290
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 89 VCEGSRFSAVMIGLAAAGGILLLLLAGGGFALYRSRRGPARKGE 132
+C+ S +M+ AAAG +L L++A G F + R R ++G+
Sbjct 207 LCQACTNSLIMLISAAAGSLLFLVVAIGIFCICRKHRKTHQEGK 250
> xla:735145 sgol1, MGC115705, sgo1; shugoshin-like 1; K11580
shugoshin-like 1
Length=663
Score = 31.2 bits (69), Expect = 2.2, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 122 RSRRGPARKGEEATRSDYVQEEAAANRKKQRQSDLVQQAEPSFWEEAE 169
RS+R ARK A +A AN KK ++ L +++E SF E AE
Sbjct 469 RSKRNTARKNSIAENELMSDVQAEANEKKITRNGLKRKSENSFTESAE 516
> dre:792854 starch binding domain 1-like
Length=533
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 3/59 (5%)
Query 100 IGLAAAGGILLLLLAGGGFALYRSRRGPARKGEEATRSDYVQ---EEAAANRKKQRQSD 155
G A GI+ +L F +YRS RG K +E + EE+A R++ + D
Sbjct 24 FGSVVAVGIIAMLSVCAAFFIYRSFRGQRGKTDEINGKETTVATGEESAVRRRRTEKDD 82
> mmu:11899 Astn1, Astn, GC14, mKIAA0289; astrotactin 1
Length=1302
Score = 29.6 bits (65), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 26/48 (54%), Gaps = 9/48 (18%)
Query 57 PCDPSTCT-ITLTSCVNGALVSKEVACP--------AEESNVCEGSRF 95
P + +T T I++TSCV G + S V+CP E + +GSRF
Sbjct 380 PVNKTTLTLISVTSCVIGLVCSSHVSCPLVVKITLHVPEHLIADGSRF 427
> hsa:460 ASTN1, ASTN, KIAA1747; astrotactin 1
Length=1294
Score = 29.3 bits (64), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 25/48 (52%), Gaps = 9/48 (18%)
Query 57 PCDPSTCT-ITLTSCVNGALVSKEVACP--------AEESNVCEGSRF 95
P + +T T I++TSCV G + S V CP E + +GSRF
Sbjct 380 PVNKTTLTLISITSCVIGLVCSSHVNCPLVVKITLHVPEHLIADGSRF 427
> ath:AT3G49850 TRB3; TRB3 (TELOMERE REPEAT BINDING FACTOR 3);
DNA binding / double-stranded telomeric DNA binding / protein
homodimerization/ single-stranded telomeric DNA binding /
transcription factor; K09422 myb proto-oncogene protein, plant
Length=295
Score = 29.3 bits (64), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 0/65 (0%)
Query 32 VAYTALGGLDEEAAPCTDSFPPTSTPCDPSTCTITLTSCVNGALVSKEVACPAEESNVCE 91
++ TAL G ++A P + + D + IT+ S NG + +++ P+ + CE
Sbjct 56 ISVTALWGSRKKAKLALKRTPLSGSRQDDNATAITIVSLANGDVGGQQIDAPSPPAGSCE 115
Query 92 GSRFS 96
R S
Sbjct 116 PPRPS 120
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 28.9 bits (63), Expect = 9.9, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 1 EGVNYGSYYPSSKECWLSDEVPNCLVPMDGAVAYTAL 37
+G N+G+Y +++C + + P CL+ +A TAL
Sbjct 395 KGYNWGNYNTETQKCEIFNVKPTCLINNSSYIATTAL 431
Lambda K H
0.312 0.129 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5235419772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40