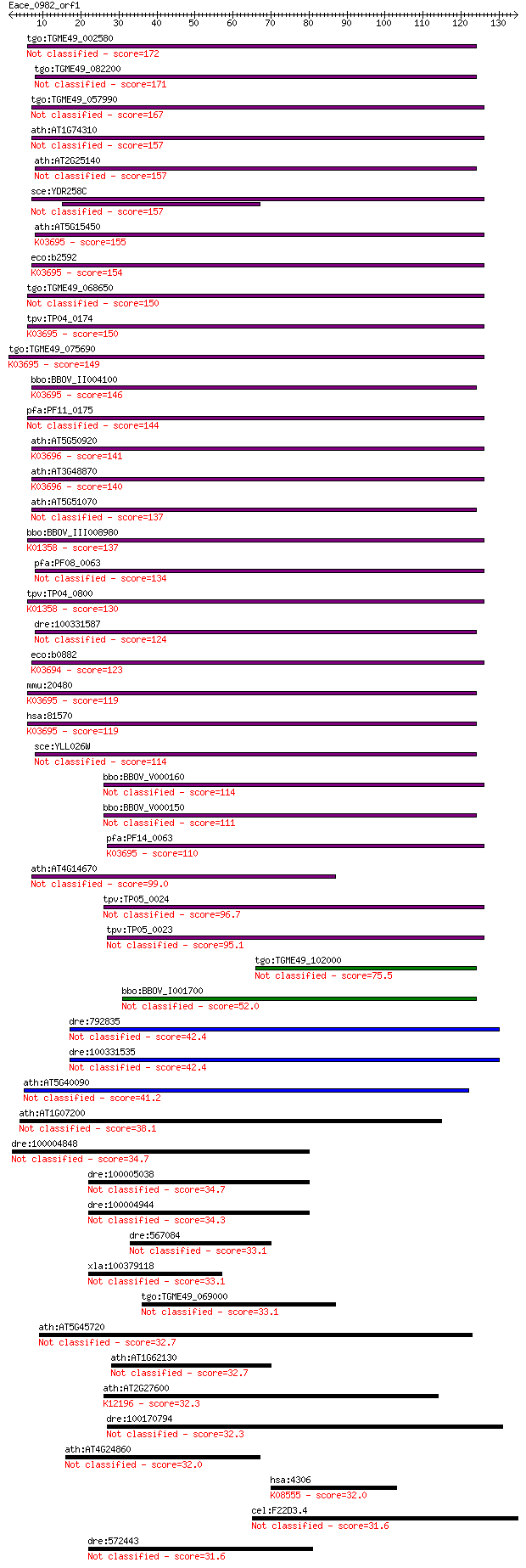
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0982_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 172 3e-43
tgo:TGME49_082200 clpB protein, putative 171 7e-43
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 167 1e-41
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 157 9e-39
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 157 9e-39
sce:YDR258C HSP78; Hsp78p 157 1e-38
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 155 5e-38
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 154 7e-38
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 150 8e-37
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 150 1e-36
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 149 2e-36
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 146 2e-35
pfa:PF11_0175 heat shock protein 101, putative 144 8e-35
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 141 5e-34
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 140 2e-33
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 137 7e-33
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 137 1e-32
pfa:PF08_0063 ClpB protein, putative 134 7e-32
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 130 1e-30
dre:100331587 suppressor of K+ transport defect 3-like 124 7e-29
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 123 1e-28
mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase B ... 119 3e-27
hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic pepti... 119 3e-27
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 114 6e-26
bbo:BBOV_V000160 clpC 114 1e-25
bbo:BBOV_V000150 clpC 111 5e-25
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 110 1e-24
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 99.0 4e-21
tpv:TP05_0024 clpC; molecular chaperone 96.7 2e-20
tpv:TP05_0023 clpC; molecular chaperone 95.1 5e-20
tgo:TGME49_102000 chaperone clpB protein, putative 75.5 4e-14
bbo:BBOV_I001700 19.m02115; chaperone clpB 52.0 5e-07
dre:792835 torsin family 3, member A 42.4 4e-04
dre:100331535 torsin family 3, member A-like 42.4 4e-04
ath:AT5G40090 ATP binding / nucleoside-triphosphatase/ nucleot... 41.2 8e-04
ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related 38.1 0.007
dre:100004848 tripartite motif protein TRIM4-like 34.7 0.086
dre:100005038 hypothetical LOC100005038 34.7 0.092
dre:100004944 ret finger protein 2-like 34.3 0.11
dre:567084 MGC152698, MGC173525; zgc:152698 33.1 0.22
xla:100379118 Ras-dva-2 small GTPase 33.1 0.23
tgo:TGME49_069000 ABC transporter, putative (EC:3.6.3.44) 33.1 0.25
ath:AT5G45720 ATP binding / DNA binding / DNA-directed DNA pol... 32.7 0.29
ath:AT1G62130 AAA-type ATPase family protein 32.7 0.35
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 32.3 0.39
dre:100170794 zgc:194342 32.3 0.42
ath:AT4G24860 AAA-type ATPase family protein 32.0 0.57
hsa:4306 NR3C2, FLJ41052, MCR, MGC133092, MLR, MR, NR3C2VIT; n... 32.0 0.59
cel:F22D3.4 hypothetical protein 31.6 0.62
dre:572443 ret finger protein-like 31.6 0.72
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 172 bits (435), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 77/118 (65%), Positives = 100/118 (84%), Gaps = 0/118 (0%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLD 65
++AV+AVA+A+ RAGL+ +N P+GTF+FLGS+GVGKTEL KA+ +F EKNLIRLD
Sbjct 604 EEAVEAVANAIIRSRAGLARRNAPIGTFLFLGSTGVGKTELAKALTAAIFHDEKNLIRLD 663
Query 66 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
M EY E H+++RL+G PPGY+ +DEGGQLTEAVRQ+PHSVVLFDE+ENAH N+++ LL
Sbjct 664 MSEYSEPHTVARLVGSPPGYVSHDEGGQLTEAVRQRPHSVVLFDEIENAHPNVFAYLL 721
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 171 bits (432), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 78/116 (67%), Positives = 98/116 (84%), Gaps = 0/116 (0%)
Query 8 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMV 67
AVKAVADA+ RAGLS + P+G+F+FLG +GVGKTEL KA+A EMF SEKNLIR+DM
Sbjct 593 AVKAVADAMVRARAGLSREGMPVGSFLFLGPTGVGKTELAKALAMEMFHSEKNLIRIDMS 652
Query 68 EYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
E+ EAHS+SRLIG PPGY+G+D GGQLTEAVR++PHSVVLFDE+E H+ + +++L
Sbjct 653 EFSEAHSVSRLIGSPPGYVGHDAGGQLTEAVRRRPHSVVLFDEIEKGHQQILNIML 708
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 167 bits (422), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 76/119 (63%), Positives = 96/119 (80%), Gaps = 0/119 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
QAV+AV A+ AGLS +N+P+G+F+FLG +GVGKTEL K VAE +FDS++ L+R DM
Sbjct 577 QAVEAVTQAILRSAAGLSRRNRPIGSFLFLGPTGVGKTELCKRVAESLFDSKERLVRFDM 636
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
EY E HS+SRLIG PPGY+G+DEGGQLTE +R+ P+SVVLFDEVE AH +W+VLL V
Sbjct 637 SEYMEQHSVSRLIGAPPGYVGHDEGGQLTEEIRRNPYSVVLFDEVEKAHSQVWNVLLQV 695
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 157 bits (397), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 72/119 (60%), Positives = 95/119 (79%), Gaps = 0/119 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
QAV AV++A+ RAGL +P G+F+FLG +GVGKTEL KA+AE++FD E L+R+DM
Sbjct 576 QAVNAVSEAILRSRAGLGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDM 635
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
EY E HS+SRLIG PPGY+G++EGGQLTEAVR++P+ V+LFDEVE AH +++ LL V
Sbjct 636 SEYMEQHSVSRLIGAPPGYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQV 694
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 157 bits (397), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 69/116 (59%), Positives = 96/116 (82%), Gaps = 0/116 (0%)
Query 8 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMV 67
AVK+VADA+ RAGLS N+P+ +FMF+G +GVGKTEL KA+A +F++E ++R+DM
Sbjct 661 AVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMS 720
Query 68 EYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
EY E HS+SRL+G PPGY+G +EGGQLTE VR++P+SVVLFDE+E AH +++++LL
Sbjct 721 EYMEKHSVSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFNILL 776
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 69/119 (57%), Positives = 96/119 (80%), Gaps = 0/119 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+A+ A++DA+ +QRAGL+ + +P+ +FMFLG +G GKTEL KA+AE +FD E N+IR DM
Sbjct 511 EAIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNVIRFDM 570
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
E+QE H++SRLIG PPGY+ ++ GGQLTEAVR+KP++VVLFDE E AH ++ +LL V
Sbjct 571 SEFQEKHTVSRLIGAPPGYVLSESGGQLTEAVRRKPYAVVLFDEFEKAHPDVSKLLLQV 629
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 32/59 (54%), Gaps = 11/59 (18%)
Query 15 ALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELV-----KAVAEEMFDS--EKNLIRLDM 66
A AIQ KN P +G +GVGKT L+ + VA E+ DS +K+L+ LD+
Sbjct 125 ARAIQILSRRTKNNP----CLIGRAGVGKTALIDGLAQRIVAGEVPDSLKDKDLVALDL 179
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 155 bits (391), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 68/118 (57%), Positives = 95/118 (80%), Gaps = 0/118 (0%)
Query 8 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMV 67
AV AVA+A+ RAGLS +P+ +FMF+G +GVGKTEL KA+A MF++E+ L+R+DM
Sbjct 656 AVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMS 715
Query 68 EYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
EY E H++SRLIG PPGY+G +EGGQLTE VR++P+SV+LFDE+E AH ++++V L +
Sbjct 716 EYMEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQI 773
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 154 bits (389), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 68/119 (57%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+AV AV++A+ RAGL+ N+P+G+F+FLG +GVGKTEL KA+A MFDS++ ++R+DM
Sbjct 575 EAVDAVSNAIRRSRAGLADPNRPIGSFLFLGPTGVGKTELCKALANFMFDSDEAMVRIDM 634
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
E+ E HS+SRL+G PPGY+G +EGG LTEAVR++P+SV+L DEVE AH +++++LL V
Sbjct 635 SEFMEKHSVSRLVGAPPGYVGYEEGGYLTEAVRRRPYSVILLDEVEKAHPDVFNILLQV 693
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 150 bits (380), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 95/120 (79%), Gaps = 0/120 (0%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLD 65
++ V++VA+A+ RAGL N+P+ + +FLG +GVGKTEL KA+A ++FD+E+ L+R D
Sbjct 600 EEGVRSVAEAIQRSRAGLCDPNRPIASLVFLGPTGVGKTELCKALARQLFDTEEALLRFD 659
Query 66 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
M EY E HS +RLIG PPGY+G D+GGQLTEAVR++P+SV+LFDE+E AH ++++ L +
Sbjct 660 MSEYMEKHSTARLIGAPPGYIGFDKGGQLTEAVRRRPYSVLLFDEMEKAHPEVFNIFLQI 719
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 150 bits (378), Expect = 1e-36, Method: Composition-based stats.
Identities = 65/120 (54%), Positives = 95/120 (79%), Gaps = 0/120 (0%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLD 65
Q+A+ AV +A+ R G++ KP+ MFLG +GVGKTEL KA+AE++FDSE+ +IR D
Sbjct 700 QEAIDAVVNAVQRSRVGMNDPKKPIAALMFLGPTGVGKTELSKAIAEQLFDSEEAIIRFD 759
Query 66 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
M EY E HS+S+L+G PPGY+G ++GG LTEA+R+KP+S++LFDE+E AH +++++LL V
Sbjct 760 MSEYMEKHSVSKLVGAPPGYIGYEQGGLLTEAIRRKPYSILLFDEIEKAHSDVYNILLQV 819
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 68/127 (53%), Positives = 100/127 (78%), Gaps = 2/127 (1%)
Query 1 NRRWLQQ--AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSE 58
+RR + Q AV+ VA+A+ RAGL+ N+P+ + FLG +GVGKTEL +++AE MFDSE
Sbjct 725 HRRVVGQDHAVQVVAEAIQRSRAGLNDPNRPIASLFFLGPTGVGKTELCRSLAELMFDSE 784
Query 59 KNLIRLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNL 118
++++DM EY E H+ISRL+G PPGY+G ++GGQLT+ VR+KP+SV+LFDE+E AH ++
Sbjct 785 DAMVKIDMSEYMEKHTISRLLGAPPGYVGYEQGGQLTDEVRKKPYSVILFDEMEKAHPDV 844
Query 119 WSVLLPV 125
++VLL +
Sbjct 845 FNVLLQI 851
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 62/117 (52%), Positives = 91/117 (77%), Gaps = 0/117 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+AV V A+ R G++ +P+ MFLG +GVGKTEL KA+AE++FD+++ +IR DM
Sbjct 646 EAVDIVTRAVQRSRVGMNDPKRPIAGLMFLGPTGVGKTELCKAIAEQLFDTDEAIIRFDM 705
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
EY E HS+SRL+G PPGY+G D+GG LTEAVR++P+S+VLFDE+E AH ++++++L
Sbjct 706 SEYMEKHSVSRLVGAPPGYIGYDQGGLLTEAVRRRPYSIVLFDEIEKAHPDVFNIML 762
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 144 bits (363), Expect = 8e-35, Method: Composition-based stats.
Identities = 65/120 (54%), Positives = 93/120 (77%), Gaps = 0/120 (0%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLD 65
+ +K+++DA+ G+ KP+GTF+FLG +GVGKTEL K +A E+F+S+ NLIR++
Sbjct 607 EDIIKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFNSKDNLIRVN 666
Query 66 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
M E+ EAHS+S++ G PPGY+G + GQLTEAVR+KPHSVVLFDE+E AH +++ VLL +
Sbjct 667 MSEFTEAHSVSKITGSPPGYVGFSDSGQLTEAVREKPHSVVLFDELEKAHADVFKVLLQI 726
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 141 bits (356), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 63/119 (52%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+AVKA++ A+ R GL N+P+ +F+F G +GVGK+EL KA+A F SE+ +IRLDM
Sbjct 615 EAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDM 674
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
E+ E H++S+LIG PPGY+G EGGQLTEAVR++P++VVLFDE+E AH ++++++L +
Sbjct 675 SEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQI 733
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 62/119 (52%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+AVKA++ A+ R GL N+P+ +F+F G +GVGK+EL KA+A F SE+ +IRLDM
Sbjct 636 EAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDM 695
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
E+ E H++S+LIG PPGY+G EGGQLTEAVR++P+++VLFDE+E AH ++++++L +
Sbjct 696 SEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNMMLQI 754
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 137 bits (346), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 60/117 (51%), Positives = 89/117 (76%), Gaps = 0/117 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+AV A++ A+ R GL ++P+ +F G +GVGKTEL KA+A F SE++++RLDM
Sbjct 634 EAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGSEESMLRLDM 693
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
EY E H++S+LIG PPGY+G +EGG LTEA+R++P +VVLFDE+E AH +++++LL
Sbjct 694 SEYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPDIFNILL 750
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 65/122 (53%), Positives = 93/122 (76%), Gaps = 5/122 (4%)
Query 6 QQAVKAVADALAIQRAGLSPKN--KPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIR 63
++AVK +A A I+RA + KN +P+G+F+F G GVGK+E+ K + + MF +E NLI+
Sbjct 665 EEAVKNIAKA--IRRAKTNIKNPERPIGSFLFCGPPGVGKSEIAKTLTKLMF-TEDNLIK 721
Query 64 LDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
LDM EY E HSISR++G PPGY G+D GGQLTE +R+ P+SVV+FDE+E AH+N+ ++LL
Sbjct 722 LDMSEYNEPHSISRILGSPPGYKGHDSGGQLTEKLRKNPYSVVMFDEIEKAHRNVLNILL 781
Query 124 PV 125
+
Sbjct 782 QI 783
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 134 bits (337), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 60/118 (50%), Positives = 88/118 (74%), Gaps = 0/118 (0%)
Query 8 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMV 67
AVK V A+ R G++ +P+ + MFLG +GVGKTEL K +A+ +FD+ + +I DM
Sbjct 787 AVKVVTKAVQRSRVGMNNPKRPIASLMFLGPTGVGKTELSKVLADVLFDTPEAVIHFDMS 846
Query 68 EYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
EY E HSIS+LIG PGY+G ++GG LT+AVR+KP+S++LFDE+E AH +++++LL V
Sbjct 847 EYMEKHSISKLIGAAPGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRV 904
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 130 bits (327), Expect = 1e-30, Method: Composition-based stats.
Identities = 59/120 (49%), Positives = 87/120 (72%), Gaps = 1/120 (0%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLD 65
++AVK V A+ + + N+P+G+F+F G GVGK+E+ +A+ + +F E NLIR+D
Sbjct 579 EEAVKNVCKAIRRAKTNIKNPNRPIGSFLFCGPPGVGKSEVARALTKYLFAKE-NLIRID 637
Query 66 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
M EY E HSISR++G PPGY G+D GGQLTE V+ P+SVV+FDE+E AH ++ ++LL +
Sbjct 638 MSEYTEPHSISRILGSPPGYKGHDTGGQLTEKVKSNPYSVVMFDEIEKAHHDVLNILLQI 697
> dre:100331587 suppressor of K+ transport defect 3-like
Length=409
Score = 124 bits (312), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 61/117 (52%), Positives = 83/117 (70%), Gaps = 2/117 (1%)
Query 8 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMF-DSEKNLIRLDM 66
A+ VA A+ + G + PL F+FLGSSG+GKTEL K VA M D +K IR+DM
Sbjct 64 AINTVASAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQVARYMHKDIKKGFIRMDM 122
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
E+QE H +++ IG PPGY+G+DEGGQLT+ ++Q P +VVLFDEVE AH ++ +V+L
Sbjct 123 SEFQEKHEVAKFIGSPPGYVGHDEGGQLTKQLKQSPSAVVLFDEVEKAHPDVLTVML 179
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 123 bits (309), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 56/119 (47%), Positives = 89/119 (74%), Gaps = 3/119 (2%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+A++A+ +A+ + RAGL ++KP+G+F+F G +GVGKTE+ +++ + L+R DM
Sbjct 465 KAIEALTEAIKMARAGLGHEHKPVGSFLFAGPTGVGKTEVTVQLSKAL---GIELLRFDM 521
Query 67 VEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
EY E H++SRLIG PPGY+G D+GG LT+AV + PH+V+L DE+E AH +++++LL V
Sbjct 522 SEYMERHTVSRLIGAPPGYVGFDQGGLLTDAVIKHPHAVLLLDEIEKAHPDVFNILLQV 580
> mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=677
Score = 119 bits (298), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 57/119 (47%), Positives = 85/119 (71%), Gaps = 2/119 (1%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMF-DSEKNLIRL 64
+ A+ V A+ + G + PL F+FLGSSG+GKTEL K A+ M D++K IRL
Sbjct 321 ESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKDAKKGFIRL 379
Query 65 DMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
DM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH ++ +++L
Sbjct 380 DMSEFQERHEVAKFIGSPPGYIGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLTIML 438
> hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease
ATP-binding subunit ClpB
Length=707
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 57/119 (47%), Positives = 85/119 (71%), Gaps = 2/119 (1%)
Query 6 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMF-DSEKNLIRL 64
+ A+ V A+ + G + PL F+FLGSSG+GKTEL K A+ M D++K IRL
Sbjct 351 ESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKDAKKGFIRL 409
Query 65 DMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
DM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH ++ +++L
Sbjct 410 DMSEFQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLTIML 468
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 56/116 (48%), Positives = 84/116 (72%), Gaps = 1/116 (0%)
Query 8 AVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMV 67
A+KAV++A+ + R+GL+ +P +F+FLG SG GKTEL K VA +F+ E +IR+D
Sbjct 586 AIKAVSNAVRLSRSGLANPRQP-ASFLFLGLSGSGKTELAKKVAGFLFNDEDMMIRVDCS 644
Query 68 EYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
E E +++S+L+G GY+G DEGG LT ++ KP+SV+LFDEVE AH ++ +V+L
Sbjct 645 ELSEKYAVSKLLGTTAGYVGYDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLTVML 700
> bbo:BBOV_V000160 clpC
Length=551
Score = 114 bits (284), Expect = 1e-25, Method: Composition-based stats.
Identities = 47/100 (47%), Positives = 71/100 (71%), Gaps = 0/100 (0%)
Query 26 KNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGY 85
K KP+G+F+ G SG GKTE+VK + ++ S+ NL++ DM EY+E+HS+S+LIG PPGY
Sbjct 299 KIKPIGSFLLCGPSGTGKTEIVKLITNYLYKSQTNLLQFDMSEYKESHSVSKLIGAPPGY 358
Query 86 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
+G++ GG L + +VLFDE+E A KN++S+ L +
Sbjct 359 VGHESGGNLINKINSVESPIVLFDEIEKADKNIFSIFLQI 398
> bbo:BBOV_V000150 clpC
Length=541
Score = 111 bits (278), Expect = 5e-25, Method: Composition-based stats.
Identities = 49/98 (50%), Positives = 67/98 (68%), Gaps = 0/98 (0%)
Query 26 KNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGY 85
K KPL +F+F G SG GKTEL K +F S K LI+L+M EY EAHSIS+++G PPGY
Sbjct 274 KTKPLSSFLFCGPSGAGKTELAKIFTYSLFKSTKQLIKLNMSEYMEAHSISKILGSPPGY 333
Query 86 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
+G +E V+ P+ V+LFDE+E AHK++ ++L
Sbjct 334 VGYNENNDFINKVKSMPNCVILFDEIEKAHKSINDLML 371
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 110 bits (274), Expect = 1e-24, Method: Composition-based stats.
Identities = 50/99 (50%), Positives = 73/99 (73%), Gaps = 1/99 (1%)
Query 27 NKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 86
NKP+GT + GSSGVGKT + +++ +F+ E NLI ++M EY + HS+S+L G PGY+
Sbjct 958 NKPIGTLLLCGSSGVGKTLCAQVISKYLFN-EDNLIVINMSEYIDKHSVSKLFGSYPGYV 1016
Query 87 GNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
G EGG+LTE+V++KP S++LFDE+E AH + VLL +
Sbjct 1017 GYKEGGELTESVKKKPFSIILFDEIEKAHSEVLHVLLQI 1055
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 46/80 (57%), Positives = 61/80 (76%), Gaps = 0/80 (0%)
Query 7 QAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
+AVKAVA A+ R GL +P G+F+FLG +GVGKTEL KA+AE++FDSE L+RLDM
Sbjct 541 EAVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDM 600
Query 67 VEYQEAHSISRLIGPPPGYM 86
EY + S+++LIG PPGY+
Sbjct 601 SEYNDKFSVNKLIGAPPGYV 620
> tpv:TP05_0024 clpC; molecular chaperone
Length=529
Score = 96.7 bits (239), Expect = 2e-20, Method: Composition-based stats.
Identities = 43/100 (43%), Positives = 65/100 (65%), Gaps = 0/100 (0%)
Query 26 KNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGY 85
+ KPLG+++ G SG GKTEL K + +F+S NLI+++M EY E H++S+LIG PPGY
Sbjct 267 ETKPLGSWLLCGPSGTGKTELAKILCYTIFNSHDNLIKINMAEYVEKHAVSKLIGSPPGY 326
Query 86 MGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
G E L+ + V+LFDE+E AH ++ ++L +
Sbjct 327 SGYGEDTILSTKFKTGSSFVILFDEIEKAHTSITDLMLQI 366
> tpv:TP05_0023 clpC; molecular chaperone
Length=502
Score = 95.1 bits (235), Expect = 5e-20, Method: Composition-based stats.
Identities = 44/102 (43%), Positives = 66/102 (64%), Gaps = 3/102 (2%)
Query 27 NKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYM 86
NKP+ F+ G SG GKT++ K ++ + ++NLI+LDM E E HS+SRL+G PPGY+
Sbjct 255 NKPIANFLLCGPSGTGKTDICKILSTYLGSEKQNLIKLDMSELAEEHSVSRLLGSPPGYV 314
Query 87 GNDEGGQ---LTEAVRQKPHSVVLFDEVENAHKNLWSVLLPV 125
G + + L + + KP+SVVL DE+E A+K L + L +
Sbjct 315 GGGKSKKSKTLVDEIIDKPNSVVLLDEIEKAYKRLCYIFLQI 356
> tgo:TGME49_102000 chaperone clpB protein, putative
Length=240
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 46/58 (79%), Gaps = 0/58 (0%)
Query 66 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
M E+ E HS+S+LIG PPGY+G + G LTEA+ +KP +V+LFDE+E AHK++ +++L
Sbjct 1 MSEFMEKHSLSKLIGAPPGYVGYGKSGLLTEAIAKKPFTVLLFDEIEKAHKDINNLML 58
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query 31 GTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGYMGNDE 90
++G GVGK L+ V+ + K + ++V +++ + L+G PPGY+G+ E
Sbjct 598 AALYYVGPPGVGKRLLLNHVSRMFGMALKYICGSNLVS---SNATNILVGSPPGYIGHRE 654
Query 91 GGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 123
GG L E ++ P+ +V F++ H N+ ++L+
Sbjct 655 GGMLCEWIKDHPYGIVAFEDAHLLHVNVVNLLI 687
> dre:792835 torsin family 3, member A
Length=367
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 58/120 (48%), Gaps = 18/120 (15%)
Query 17 AIQRAGLSPK-NKPLGTFMFLGSSGVGKTELVKAVAEEMFDS--EKNLIRLDMVEYQEAH 73
AIQ +P+ NKPL T F G SG GK + + VA+ ++ + +RL + + H
Sbjct 119 AIQGFIKNPESNKPL-TLSFHGWSGTGKNFVARIVADNLYRDGIKSECVRLFIAPFHFPH 177
Query 74 SISRLIGPPPGYMGNDEGGQLTEAVR----QKPHSVVLFDEVENAHKNLWSVLLPVRKYF 129
+RL+ + GQL EA+R + P ++ +FDE E H L + P ++
Sbjct 178 --ARLV--------DVYKGQLREAIRDMVLRCPQTLFIFDEAEKLHPGLIDAIKPYMDHY 227
> dre:100331535 torsin family 3, member A-like
Length=367
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 58/120 (48%), Gaps = 18/120 (15%)
Query 17 AIQRAGLSPK-NKPLGTFMFLGSSGVGKTELVKAVAEEMFDS--EKNLIRLDMVEYQEAH 73
AIQ +P+ NKPL T F G SG GK + + VA+ ++ + +RL + + H
Sbjct 119 AIQGFIKNPESNKPL-TLSFHGWSGTGKNFVARIVADNLYRDGIKSECVRLFIAPFHFPH 177
Query 74 SISRLIGPPPGYMGNDEGGQLTEAVR----QKPHSVVLFDEVENAHKNLWSVLLPVRKYF 129
+RL+ + GQL EA+R + P ++ +FDE E H L + P ++
Sbjct 178 --ARLV--------DVYKGQLREAIRDMVLRCPQTLFIFDEAEKLHPGLIDAIKPYMDHY 227
> ath:AT5G40090 ATP binding / nucleoside-triphosphatase/ nucleotide
binding / transmembrane receptor
Length=459
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 59/120 (49%), Gaps = 10/120 (8%)
Query 5 LQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRL 64
+ + +K V D LA++ NK + T GS+GVGKT L + + E+F + + + L
Sbjct 184 MDRHMKVVYDLLALE------VNKEVRTIGIWGSAGVGKTTLARYIYAEIFVNFQTHVFL 237
Query 65 DMVEYQEAHSISRLIG---PPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSV 121
D VE + + + G P +G ++TEA R+ +++ D+V N + W +
Sbjct 238 DNVENMK-DKLLKFEGEEDPTVIISSYHDGHEITEARRKHRKILLIADDVNNMEQGKWII 296
> ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related
Length=979
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 51/112 (45%), Gaps = 9/112 (8%)
Query 4 WLQQAVKAVADALAIQRAGLSPKNKPLGTFM-FLGSSGVGKTELVKAVAEEMFDSEKNLI 62
W +AV A++ + + + +N+ G ++ LG VGK ++ ++E F + N I
Sbjct 631 WQTEAVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSEVFFGGKVNYI 690
Query 63 RLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENA 114
V++ H + G +T + +KPHSVVL + VE A
Sbjct 691 ---CVDFGAEH-----CSLDDKFRGKTVVDYVTGELSRKPHSVVLLENVEKA 734
> dre:100004848 tripartite motif protein TRIM4-like
Length=621
Score = 34.7 bits (78), Expect = 0.086, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 41/85 (48%), Gaps = 7/85 (8%)
Query 2 RRWLQQAVKAVADALAIQRA--GLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEK 59
R LQ++ +A + ++ G KNKP T + +G +G GKT L+ + M ++
Sbjct 112 RYRLQRSTDNLAQSELFRKVTFGERDKNKPHKTILIVGETGTGKTTLINVMVNYMLGVKR 171
Query 60 N-----LIRLDMVEYQEAHSISRLI 79
I D ++ +AHS + +I
Sbjct 172 EDKVWFEITDDQSDWDQAHSQTSII 196
> dre:100005038 hypothetical LOC100005038
Length=498
Score = 34.7 bits (78), Expect = 0.092, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Query 22 GLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKN-----LIRLDMVEYQEAHSIS 76
G KNKP T + +G +G GKT+L+ + M E+ I D + AHS +
Sbjct 50 GKRDKNKPHKTILMVGETGTGKTKLINTMVNYMLGVEREDKVWFEITDDQSDRSSAHSQT 109
Query 77 RLI 79
+I
Sbjct 110 SII 112
> dre:100004944 ret finger protein 2-like
Length=600
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Query 22 GLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKN-----LIRLDMVEYQEAHSIS 76
G +NKP T + +G +GVGKT LV + M E+ I D + AHS +
Sbjct 131 GKRDENKPHKTILIVGETGVGKTTLVNVMVNYMLGVEREDKVWFEITDDQSDQSSAHSQT 190
Query 77 RLI 79
+I
Sbjct 191 SII 193
> dre:567084 MGC152698, MGC173525; zgc:152698
Length=211
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 26/39 (66%), Gaps = 2/39 (5%)
Query 33 FMFLGSSGVGKTELVKAVAEEMFDSE--KNLIRLDMVEY 69
F+FLG++GVGKT L+ ++ FDS+ + + L +EY
Sbjct 13 FVFLGAAGVGKTALITRFLQDRFDSKYTRTVEELHALEY 51
> xla:100379118 Ras-dva-2 small GTPase
Length=209
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 22 GLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFD 56
LS K K +FLG++GVGKT L++ ++ FD
Sbjct 2 SLSTKEKRQIRLVFLGAAGVGKTSLIRRFLQDTFD 36
> tgo:TGME49_069000 ABC transporter, putative (EC:3.6.3.44)
Length=1056
Score = 33.1 bits (74), Expect = 0.25, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 29/53 (54%), Gaps = 3/53 (5%)
Query 36 LGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQE--AHSISRLIGPPPGYM 86
+G SGVGK+ L+K + MFD +R+D + +E HS R IG P M
Sbjct 835 VGPSGVGKSTLIKLLF-RMFDPASGTVRIDDQDVKELDLHSFRRQIGVVPQDM 886
> ath:AT5G45720 ATP binding / DNA binding / DNA-directed DNA polymerase/
nucleoside-triphosphatase/ nucleotide binding
Length=966
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 27/120 (22%), Positives = 53/120 (44%), Gaps = 14/120 (11%)
Query 9 VKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMF-----DSEKNLIR 63
V+A+++A+A +R GL ++F G +G GKT + A + S+ +
Sbjct 364 VQALSNAIAKRRVGL--------LYVFHGPNGTGKTSCARVFARALNCHSTEQSKPCGVC 415
Query 64 LDMVEYQEAHS-ISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVL 122
V Y + + R +GP + + + +QK V++FD+ + + W+ L
Sbjct 416 SSCVSYDDGKNRYIREMGPVKSFDFENLLDKTNIRQQQKQQLVLIFDDCDTMSTDCWNTL 475
> ath:AT1G62130 AAA-type ATPase family protein
Length=1025
Score = 32.7 bits (73), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query 28 KPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEY 69
KP + G SG GKT L KAVA E + NLI + M +
Sbjct 768 KPCNGILLFGPSGTGKTMLAKAVATE---AGANLINMSMSRW 806
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 32.3 bits (72), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 40/88 (45%), Gaps = 13/88 (14%)
Query 26 KNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDMVEYQEAHSISRLIGPPPGY 85
K +P F+ G G GK+ L KAVA E + ++ D+V S+ +G
Sbjct 161 KRRPWRAFLLYGPPGTGKSYLAKAVATEADSTFFSVSSSDLV--------SKWMGESEKL 212
Query 86 MGNDEGGQLTEAVRQKPHSVVLFDEVEN 113
+ N L E R+ S++ DE+++
Sbjct 213 VSN-----LFEMARESAPSIIFVDEIDS 235
> dre:100170794 zgc:194342
Length=323
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 45/109 (41%), Gaps = 15/109 (13%)
Query 27 NKPLGTFMFLGSSGVGKTELVKAVAEEMFDS-EKNLIRLDMVEYQEAHSISRLIGPPPGY 85
NKPL F G++G GK + K +A ++ EK+ + H+ P
Sbjct 83 NKPL-VLSFHGTAGTGKNHVAKIIARNVYKKGEKS---------KHVHTFISQFHFPHQE 132
Query 86 MGNDEGGQLTE----AVRQKPHSVVLFDEVENAHKNLWSVLLPVRKYFY 130
+ QL + V P S+ +FDE++ H L ++ P Y Y
Sbjct 133 NVHMYSAQLKQWIHGNVSSFPRSMFIFDEMDKMHPELIDIIKPFLDYNY 181
> ath:AT4G24860 AAA-type ATPase family protein
Length=1122
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 27/54 (50%), Gaps = 6/54 (11%)
Query 16 LAIQRAGLSPK---NKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKNLIRLDM 66
L +QR L K KP + G G GKT L KAVA+E ++ N I + M
Sbjct 838 LPLQRPELFCKGELTKPCKGILLFGPPGTGKTMLAKAVAKE---ADANFINISM 888
> hsa:4306 NR3C2, FLJ41052, MCR, MGC133092, MLR, MR, NR3C2VIT;
nuclear receptor subfamily 3, group C, member 2; K08555 mineralocorticoid
receptor
Length=984
Score = 32.0 bits (71), Expect = 0.59, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query 70 QEAHSISRLIGPP-PGYMGNDEGGQLTEAVRQKP 102
++ +S+S ++GPP PG+ GN EG ++Q+P
Sbjct 464 KDYYSLSGILGPPVPGFDGNCEGSGFPVGIKQEP 497
> cel:F22D3.4 hypothetical protein
Length=439
Score = 31.6 bits (70), Expect = 0.62, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 35/71 (49%), Gaps = 3/71 (4%)
Query 65 DMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVL-FDEVENAHKNLWSVLL 123
D+VE Q A S +R +G P Y ++G + E V FDE A + + +L+
Sbjct 139 DIVEDQNAQSSARSLGKP--YYYEEDGFLVIENANVYSQGVYFCFDEDSVASQRFFYILI 196
Query 124 PVRKYFYRETK 134
P+ F+ ++K
Sbjct 197 PILPVFHIDSK 207
> dre:572443 ret finger protein-like
Length=644
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 5/64 (7%)
Query 22 GLSPKNKPLGTFMFLGSSGVGKTELVKAVAEEMFDSEKN-----LIRLDMVEYQEAHSIS 76
G KNKP T + +G +G GKT+L+ + M ++ I D AHS +
Sbjct 174 GQRDKNKPHKTILLVGETGTGKTKLINTMINYMLGVKREDKVWFEITDDQSNETSAHSQT 233
Query 77 RLIG 80
+I
Sbjct 234 SIIA 237
Lambda K H
0.317 0.134 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40