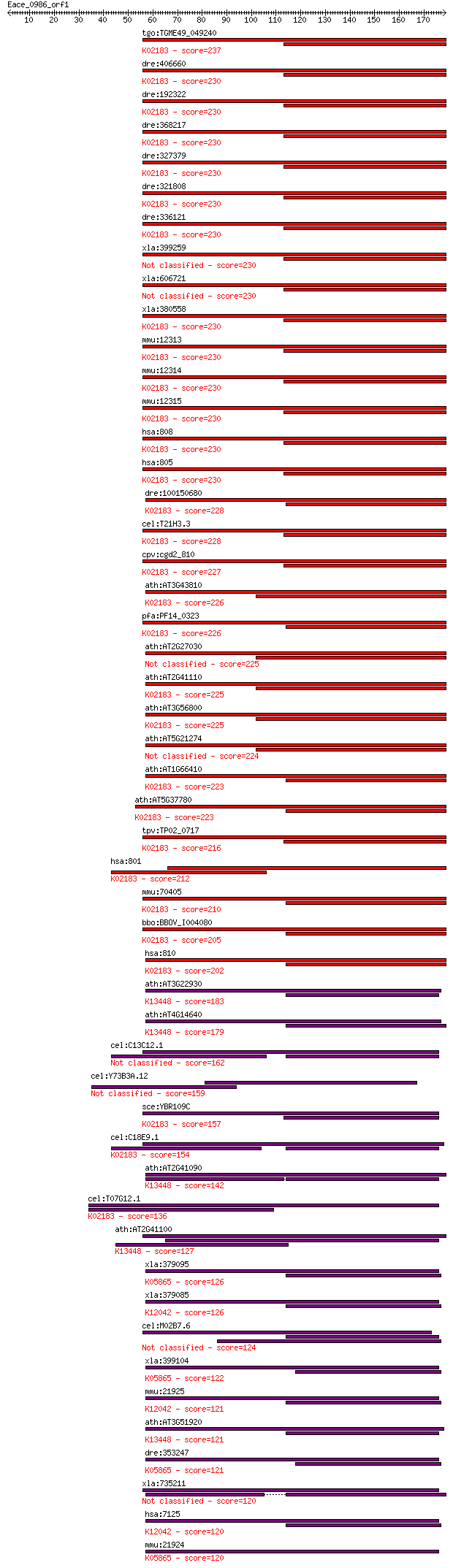
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0986_orf1
Length=178
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_049240 calmodulin ; K02183 calmodulin 237 2e-62
dre:406660 calm1a, CaMbeta, sb:cb617, wu:fb08d09, wu:fb69c08, ... 230 2e-60
dre:192322 calm2b, calm2, cb169, zgc:64036; calmodulin 2b, (ph... 230 2e-60
dre:368217 calm1b, CaMbeta-2, zgc:73137; calmodulin 1b; K02183... 230 2e-60
dre:327379 calm3a, CaMgamma, MGC192068, calm2g, wu:fi05b08, zg... 230 2e-60
dre:321808 calm3b, wu:fb36a09, zgc:55591, zgc:76987; calmoduli... 230 2e-60
dre:336121 calm2a, CaMalpha, calm2d, cb815, wu:fj49d04, zgc:73... 230 2e-60
xla:399259 calm2, calm1, cam; cam (EC:2.7.11.19) 230 2e-60
xla:606721 calm1, calm2a, calml2, cami, dd132; calmodulin 1 (p... 230 2e-60
xla:380558 calm2, MGC64460, calm1, camii, phkd, phkd2; calmodu... 230 2e-60
mmu:12313 Calm1, AI256814, AI327027, AI461935, AL024000, CaM, ... 230 2e-60
mmu:12314 Calm2, 1500001E21Rik, AL024017, Calm1, Calm3; calmod... 230 2e-60
mmu:12315 Calm3, CaMA, Calm1, Calm2, R75142; calmodulin 3 (EC:... 230 2e-60
hsa:808 CALM3, CALM1, CALM2, PHKD, PHKD3; calmodulin 3 (phosph... 230 2e-60
hsa:805 CALM2, CALM1, CALM3, CAMII, FLJ99410, PHKD, PHKD2; cal... 230 2e-60
dre:100150680 calmodulin 2-like; K02183 calmodulin 228 6e-60
cel:T21H3.3 cmd-1; CalModulin family member (cmd-1); K02183 ca... 228 7e-60
cpv:cgd2_810 calmodulin ; K02183 calmodulin 227 2e-59
ath:AT3G43810 CAM7; CAM7 (CALMODULIN 7); calcium ion binding; ... 226 3e-59
pfa:PF14_0323 calmodulin; K02183 calmodulin 226 3e-59
ath:AT2G27030 CAM5; CAM5 (CALMODULIN 5); calcium ion binding 225 8e-59
ath:AT2G41110 CAM2; CAM2 (CALMODULIN 2); calcium ion binding /... 225 8e-59
ath:AT3G56800 CAM3; CAM3 (CALMODULIN 3); calcium ion binding; ... 225 8e-59
ath:AT5G21274 CAM6; CAM6 (CALMODULIN 6); calcium ion binding 224 9e-59
ath:AT1G66410 CAM4; CAM4 (calmodulin 4); calcium ion binding /... 223 2e-58
ath:AT5G37780 CAM1; CAM1 (CALMODULIN 1); calcium ion binding; ... 223 2e-58
tpv:TP02_0717 calmodulin; K02183 calmodulin 216 5e-56
hsa:801 CALM1, CALM2, CALM3, CALML2, CAMI, DD132, PHKD; calmod... 212 5e-55
mmu:70405 Calml3, 2310068O22Rik, AI326174; calmodulin-like 3; ... 210 2e-54
bbo:BBOV_I004080 19.m02335; calmodulin; K02183 calmodulin 205 6e-53
hsa:810 CALML3, CLP; calmodulin-like 3; K02183 calmodulin 202 4e-52
ath:AT3G22930 calmodulin, putative; K13448 calcium-binding pro... 183 2e-46
ath:AT4G14640 CAM8; CAM8 (CALMODULIN 8); calcium ion binding; ... 179 6e-45
cel:C13C12.1 cal-1; CALmodulin related genes family member (ca... 162 5e-40
cel:Y73B3A.12 hypothetical protein 159 5e-39
sce:YBR109C CMD1; Calmodulin; Ca++ binding protein that regula... 157 2e-38
cel:C18E9.1 cal-2; CALmodulin related genes family member (cal... 154 1e-37
ath:AT2G41090 calmodulin-like calcium-binding protein, 22 kDa ... 142 6e-34
cel:T07G12.1 cal-4; CALmodulin related genes family member (ca... 136 4e-32
ath:AT2G41100 TCH3; TCH3 (TOUCH 3); calcium ion binding; K1344... 127 2e-29
xla:379095 tnnc2, MGC53213; troponin C type 2 (fast); K05865 t... 126 5e-29
xla:379085 tnnc2, MGC52923; fast skeletal troponin C beta; K12... 126 5e-29
cel:M02B7.6 cal-3; CALmodulin related genes family member (cal-3) 124 2e-28
xla:399104 tnnc1, MGC80066; cardiac troponin C; K05865 troponi... 122 9e-28
mmu:21925 Tnnc2, Tncs; troponin C2, fast; K12042 troponin C, s... 121 1e-27
ath:AT3G51920 CAM9; CAM9 (CALMODULIN 9); calcium ion binding; ... 121 1e-27
dre:353247 tnnc1a, cTnC, tnnc1, zgc:103465; troponin C type 1a... 121 2e-27
xla:735211 cetn4, MGC130946; centrin 4 120 2e-27
hsa:7125 TNNC2; troponin C type 2 (fast); K12042 troponin C, s... 120 2e-27
mmu:21924 Tnnc1, AI874626, TnC, cTnC, cTnI, tncc; troponin C, ... 120 2e-27
> tgo:TGME49_049240 calmodulin ; K02183 calmodulin
Length=149
Score = 237 bits (604), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 116/123 (94%), Positives = 119/123 (96%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTD+E EL
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDTEEEL 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
IEAFKVFDRDGNG ISAAELRHVMTNLGEKLTDEEVDEMIREAD+DGDGQINYEEFV MM
Sbjct 87 IEAFKVFDRDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEFVKMM 146
Query 176 LAK 178
+AK
Sbjct 147 MAK 149
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
+M K
Sbjct 71 TLMARK 76
> dre:406660 calm1a, CaMbeta, sb:cb617, wu:fb08d09, wu:fb69c08,
wu:fj34a08, zgc:63926; calmodulin 1a; K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> dre:192322 calm2b, calm2, cb169, zgc:64036; calmodulin 2b, (phosphorylase
kinase, delta); K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> dre:368217 calm1b, CaMbeta-2, zgc:73137; calmodulin 1b; K02183
calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> dre:327379 calm3a, CaMgamma, MGC192068, calm2g, wu:fi05b08,
zgc:86728; calmodulin 3a (phosphorylase kinase, delta); K02183
calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> dre:321808 calm3b, wu:fb36a09, zgc:55591, zgc:76987; calmodulin
3b (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> dre:336121 calm2a, CaMalpha, calm2d, cb815, wu:fj49d04, zgc:73045;
calmodulin 2a (phosphorylase kinase, delta); K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> xla:399259 calm2, calm1, cam; cam (EC:2.7.11.19)
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> xla:606721 calm1, calm2a, calml2, cami, dd132; calmodulin 1
(phosphorylase kinase, delta)
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> xla:380558 calm2, MGC64460, calm1, camii, phkd, phkd2; calmodulin
2 (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> mmu:12313 Calm1, AI256814, AI327027, AI461935, AL024000, CaM,
Calm, Calm2, Calm3; calmodulin 1 (EC:2.7.11.19); K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> mmu:12314 Calm2, 1500001E21Rik, AL024017, Calm1, Calm3; calmodulin
2 (EC:2.7.11.19); K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> mmu:12315 Calm3, CaMA, Calm1, Calm2, R75142; calmodulin 3 (EC:2.7.11.19);
K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> hsa:808 CALM3, CALM1, CALM2, PHKD, PHKD3; calmodulin 3 (phosphorylase
kinase, delta) (EC:2.7.11.19); K02183 calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> hsa:805 CALM2, CALM1, CALM3, CAMII, FLJ99410, PHKD, PHKD2; calmodulin
2 (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=149
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/123 (91%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 146
Query 176 LAK 178
AK
Sbjct 147 TAK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> dre:100150680 calmodulin 2-like; K02183 calmodulin
Length=229
Score = 228 bits (582), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 112/122 (91%), Positives = 117/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 108 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEIR 167
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 168 EAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMT 227
Query 177 AK 178
AK
Sbjct 228 AK 229
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 92 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT 151
Query 174 MMLAK 178
MM K
Sbjct 152 MMARK 156
> cel:T21H3.3 cmd-1; CalModulin family member (cmd-1); K02183
calmodulin
Length=149
Score = 228 bits (582), Expect = 7e-60, Method: Compositional matrix adjust.
Identities = 111/123 (90%), Positives = 116/123 (94%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM
Sbjct 87 REAFRVFDKDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVTMM 146
Query 176 LAK 178
K
Sbjct 147 TTK 149
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
MM K
Sbjct 71 TMMARK 76
> cpv:cgd2_810 calmodulin ; K02183 calmodulin
Length=149
Score = 227 bits (578), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 111/123 (90%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
+ITTKELGTVMRSLGQNPTEAEL DMINEVDADGNGTIDFPEFL+LMARKMKDTD+E EL
Sbjct 27 SITTKELGTVMRSLGQNPTEAELLDMINEVDADGNGTIDFPEFLSLMARKMKDTDTEDEL 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
IEAFKVFDRDGNG ISAAELRHVMTNLGEKL+DEEVDEMIREAD+DGDGQI YEEF MM
Sbjct 87 IEAFKVFDRDGNGFISAAELRHVMTNLGEKLSDEEVDEMIREADVDGDGQIMYEEFTKMM 146
Query 176 LAK 178
L+K
Sbjct 147 LSK 149
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 11 AEFKEAFSLFDKDGDGSITTKELGTVMRSLGQNPTEAELLDMINEVDADGNGTIDFPEFL 70
Query 173 GMMLAK 178
+M K
Sbjct 71 SLMARK 76
> ath:AT3G43810 CAM7; CAM7 (CALMODULIN 7); calcium ion binding;
K02183 calmodulin
Length=149
Score = 226 bits (576), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 111/122 (90%), Positives = 116/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMARKMKDTDSE EL
Sbjct 28 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSEEELK 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+D NG ISAAELRHVMTNLGEKLTDEEVDEMIREAD+DGDGQINYEEFV +M+
Sbjct 88 EAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEFVKVMM 147
Query 177 AK 178
AK
Sbjct 148 AK 149
Score = 62.8 bits (151), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 102 MARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADID 161
MA ++ D D E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D D
Sbjct 1 MADQLTD-DQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDAD 59
Query 162 GDGQINYEEFVGMMLAK 178
G+G I++ EF+ +M K
Sbjct 60 GNGTIDFPEFLNLMARK 76
> pfa:PF14_0323 calmodulin; K02183 calmodulin
Length=149
Score = 226 bits (576), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 110/123 (89%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
ITTKELGTVMRSLGQNPTEAELQDMINE+D DGNGTIDFPEFLTLMARK+KDTD+E EL
Sbjct 27 TITTKELGTVMRSLGQNPTEAELQDMINEIDTDGNGTIDFPEFLTLMARKLKDTDTEEEL 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
IEAF+VFDRDG+G ISA ELRHVMTNLGEKLT+EEVDEMIREADIDGDGQINYEEFV MM
Sbjct 87 IEAFRVFDRDGDGYISADELRHVMTNLGEKLTNEEVDEMIREADIDGDGQINYEEFVKMM 146
Query 176 LAK 178
+AK
Sbjct 147 IAK 149
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEIDTDGNGTIDFPEFLT 71
Query 174 MMLAK 178
+M K
Sbjct 72 LMARK 76
> ath:AT2G27030 CAM5; CAM5 (CALMODULIN 5); calcium ion binding
Length=149
Score = 225 bits (573), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 110/122 (90%), Positives = 116/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMARKMKDTDSE EL
Sbjct 28 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSEEELK 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+D NG ISAAELRHVMTNLGEKLTDEEVDEMI+EAD+DGDGQINYEEFV +M+
Sbjct 88 EAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINYEEFVKVMM 147
Query 177 AK 178
AK
Sbjct 148 AK 149
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 102 MARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADID 161
MA ++ D D E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D D
Sbjct 1 MADQLTD-DQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDAD 59
Query 162 GDGQINYEEFVGMMLAK 178
G+G I++ EF+ +M K
Sbjct 60 GNGTIDFPEFLNLMARK 76
> ath:AT2G41110 CAM2; CAM2 (CALMODULIN 2); calcium ion binding
/ protein binding; K02183 calmodulin
Length=149
Score = 225 bits (573), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 110/122 (90%), Positives = 116/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMARKMKDTDSE EL
Sbjct 28 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSEEELK 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+D NG ISAAELRHVMTNLGEKLTDEEVDEMI+EAD+DGDGQINYEEFV +M+
Sbjct 88 EAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINYEEFVKVMM 147
Query 177 AK 178
AK
Sbjct 148 AK 149
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 102 MARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADID 161
MA ++ D D E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D D
Sbjct 1 MADQLTD-DQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDAD 59
Query 162 GDGQINYEEFVGMMLAK 178
G+G I++ EF+ +M K
Sbjct 60 GNGTIDFPEFLNLMARK 76
> ath:AT3G56800 CAM3; CAM3 (CALMODULIN 3); calcium ion binding;
K02183 calmodulin
Length=149
Score = 225 bits (573), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 110/122 (90%), Positives = 116/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMARKMKDTDSE EL
Sbjct 28 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSEEELK 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+D NG ISAAELRHVMTNLGEKLTDEEVDEMI+EAD+DGDGQINYEEFV +M+
Sbjct 88 EAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINYEEFVKVMM 147
Query 177 AK 178
AK
Sbjct 148 AK 149
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 102 MARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADID 161
MA ++ D D E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D D
Sbjct 1 MADQLTD-DQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDAD 59
Query 162 GDGQINYEEFVGMMLAK 178
G+G I++ EF+ +M K
Sbjct 60 GNGTIDFPEFLNLMARK 76
> ath:AT5G21274 CAM6; CAM6 (CALMODULIN 6); calcium ion binding
Length=149
Score = 224 bits (572), Expect = 9e-59, Method: Compositional matrix adjust.
Identities = 110/122 (90%), Positives = 116/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMARKMKDTDSE EL
Sbjct 28 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSEEELK 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+D NG ISAAELRHVMTNLGEKL+DEEVDEMIREAD+DGDGQINYEEFV +M+
Sbjct 88 EAFRVFDKDQNGFISAAELRHVMTNLGEKLSDEEVDEMIREADVDGDGQINYEEFVKVMM 147
Query 177 AK 178
AK
Sbjct 148 AK 149
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 102 MARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADID 161
MA ++ D D E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D D
Sbjct 1 MADQLTD-DQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDAD 59
Query 162 GDGQINYEEFVGMMLAK 178
G+G I++ EF+ +M K
Sbjct 60 GNGTIDFPEFLNLMARK 76
> ath:AT1G66410 CAM4; CAM4 (calmodulin 4); calcium ion binding
/ signal transducer; K02183 calmodulin
Length=149
Score = 223 bits (569), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 109/122 (89%), Positives = 116/122 (95%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMA+KMKDTDSE EL
Sbjct 28 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMAKKMKDTDSEEELK 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+D NG ISAAELRHVMTNLGEKLTDEEV+EMIREAD+DGDGQINYEEFV +M+
Sbjct 88 EAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQINYEEFVKIMM 147
Query 177 AK 178
AK
Sbjct 148 AK 149
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +MI E D DG+G I++ EF+
Sbjct 12 EFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query 174 MMLAK 178
+M K
Sbjct 72 LMAKK 76
> ath:AT5G37780 CAM1; CAM1 (CALMODULIN 1); calcium ion binding;
K02183 calmodulin
Length=164
Score = 223 bits (568), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 110/126 (87%), Positives = 117/126 (92%), Gaps = 0/126 (0%)
Query 53 FKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSE 112
F ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL LMA+KMKDTDSE
Sbjct 39 FGGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMAKKMKDTDSE 98
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
EL EAF+VFD+D NG ISAAELRHVMTNLGEKLTDEEV+EMIREAD+DGDGQINYEEFV
Sbjct 99 EELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQINYEEFV 158
Query 173 GMMLAK 178
+M+AK
Sbjct 159 KIMMAK 164
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 44/80 (55%), Gaps = 15/80 (18%)
Query 114 ELIEAFKVFDRDGN---------------GLISAAELRHVMTNLGEKLTDEEVDEMIREA 158
E EAF +FD+DG+ G I+ EL VM +LG+ T+ E+ +MI E
Sbjct 12 EFKEAFSLFDKDGDALNMCLLVANLFRFGGCITTKELGTVMRSLGQNPTEAELQDMINEV 71
Query 159 DIDGDGQINYEEFVGMMLAK 178
D DG+G I++ EF+ +M K
Sbjct 72 DADGNGTIDFPEFLNLMAKK 91
> tpv:TP02_0717 calmodulin; K02183 calmodulin
Length=149
Score = 216 bits (549), Expect = 5e-56, Method: Compositional matrix adjust.
Identities = 101/123 (82%), Positives = 117/123 (95%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
+IT+KELGT+MRSLGQNPTEAELQDMINE+DA+ NG+IDFPEFLTLMARKMK+ D+E EL
Sbjct 27 SITSKELGTIMRSLGQNPTEAELQDMINEIDANSNGSIDFPEFLTLMARKMKECDTEEEL 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
I+AFKVFDRDGNG ISA ELRHVMTNLGE+LTDEEVDEM+READ+DGDG+INYEEFV +M
Sbjct 87 IQAFKVFDRDGNGFISAQELRHVMTNLGERLTDEEVDEMLREADVDGDGKINYEEFVKLM 146
Query 176 LAK 178
++K
Sbjct 147 VSK 149
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+DG+G I++ EL +M +LG+ T+ E+ +MI E D + +G I++ EF+
Sbjct 11 AEFKEAFALFDKDGDGSITSKELGTIMRSLGQNPTEAELQDMINEIDANSNGSIDFPEFL 70
Query 173 GMMLAK 178
+M K
Sbjct 71 TLMARK 76
> hsa:801 CALM1, CALM2, CALM3, CALML2, CAMI, DD132, PHKD; calmodulin
1 (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=113
Score = 212 bits (540), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 103/113 (91%), Positives = 108/113 (95%), Gaps = 0/113 (0%)
Query 66 MRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELIEAFKVFDRD 125
MRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLT+MARKMKDTDSE E+ EAF+VFD+D
Sbjct 1 MRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKD 60
Query 126 GNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMMLAK 178
GNG ISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQ+NYEEFV MM AK
Sbjct 61 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 113
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 43 QRAARVLLRMFKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLM 102
+ A RV + I+ EL VM +LG+ T+ E+ +MI E D DG+G +++ EF+ +M
Sbjct 51 REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMM 110
Query 103 ARK 105
K
Sbjct 111 TAK 113
> mmu:70405 Calml3, 2310068O22Rik, AI326174; calmodulin-like 3;
K02183 calmodulin
Length=149
Score = 210 bits (534), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 97/123 (78%), Positives = 114/123 (92%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
+ITT+ELGTVMRSLGQNPTEAELQ M+NE+D DGNGT+DFPEFLT+M+RKMKDTDSE E+
Sbjct 27 SITTQELGTVMRSLGQNPTEAELQGMVNEIDKDGNGTVDFPEFLTMMSRKMKDTDSEEEI 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG +SAAELRHVMT LGEKL+DEEVDEMI+ AD DGDGQ+NYEEFV M+
Sbjct 87 REAFRVFDKDGNGFVSAAELRHVMTKLGEKLSDEEVDEMIQAADTDGDGQVNYEEFVHML 146
Query 176 LAK 178
++K
Sbjct 147 VSK 149
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ M+ E D DG+G +++ EF+
Sbjct 12 EFKEAFSLFDKDGDGSITTQELGTVMRSLGQNPTEAELQGMVNEIDKDGNGTVDFPEFLT 71
Query 174 MMLAK 178
MM K
Sbjct 72 MMSRK 76
> bbo:BBOV_I004080 19.m02335; calmodulin; K02183 calmodulin
Length=149
Score = 205 bits (522), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 97/123 (78%), Positives = 112/123 (91%), Gaps = 0/123 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
+ITTKELGTVMRSLGQNPTEAEL DMIN++D G G IDFPEFL LMARKMK+ D+E EL
Sbjct 27 SITTKELGTVMRSLGQNPTEAELADMINDIDTSGTGAIDFPEFLILMARKMKEGDTEEEL 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
++AFKVFDRDGNG ISA ELRHVMTNLGEKLT+EEV+EM+READ+DGDG+INYEEFV +M
Sbjct 87 VQAFKVFDRDGNGFISAQELRHVMTNLGEKLTNEEVEEMLREADVDGDGKINYEEFVKLM 146
Query 176 LAK 178
++K
Sbjct 147 ISK 149
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FDRDG+G I+ EL VM +LG+ T+ E+ +MI + D G G I++ EF+
Sbjct 12 EFKEAFSLFDRDGDGSITTKELGTVMRSLGQNPTEAELADMINDIDTSGTGAIDFPEFLI 71
Query 174 MMLAK 178
+M K
Sbjct 72 LMARK 76
> hsa:810 CALML3, CLP; calmodulin-like 3; K02183 calmodulin
Length=149
Score = 202 bits (515), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 95/122 (77%), Positives = 113/122 (92%), Gaps = 0/122 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITT+ELGTVMRSLGQNPTEAEL+DM++E+D DGNGT+DFPEFL +MARKMKDTD+E E+
Sbjct 28 ITTRELGTVMRSLGQNPTEAELRDMMSEIDRDGNGTVDFPEFLGMMARKMKDTDNEEEIR 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAF+VFD+DGNG +SAAELRHVMT LGEKL+DEEVDEMIR AD DGDGQ+NYEEFV +++
Sbjct 88 EAFRVFDKDGNGFVSAAELRHVMTRLGEKLSDEEVDEMIRAADTDGDGQVNYEEFVRVLV 147
Query 177 AK 178
+K
Sbjct 148 SK 149
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+ EL VM +LG+ T+ E+ +M+ E D DG+G +++ EF+G
Sbjct 12 EFKEAFSLFDKDGDGCITTRELGTVMRSLGQNPTEAELRDMMSEIDRDGNGTVDFPEFLG 71
Query 174 MMLAK 178
MM K
Sbjct 72 MMARK 76
> ath:AT3G22930 calmodulin, putative; K13448 calcium-binding protein
CML
Length=173
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 87/120 (72%), Positives = 105/120 (87%), Gaps = 0/120 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
IT EL TV+RSL QNPTE ELQDMI E+D+DGNGTI+F EFL LMA ++++TD++ EL
Sbjct 51 ITADELATVIRSLDQNPTEQELQDMITEIDSDGNGTIEFSEFLNLMANQLQETDADEELK 110
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAFKVFD+D NG ISA+ELRHVM NLGEKLTDEEVD+MI+EAD+DGDGQ+NY+EFV MM+
Sbjct 111 EAFKVFDKDQNGYISASELRHVMINLGEKLTDEEVDQMIKEADLDGDGQVNYDEFVRMMM 170
Score = 55.8 bits (133), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+A EL V+ +L + T++E+ +MI E D DG+G I + EF+
Sbjct 35 EFKEAFCLFDKDGDGCITADELATVIRSLDQNPTEQELQDMITEIDSDGNGTIEFSEFLN 94
Query 174 MM 175
+M
Sbjct 95 LM 96
> ath:AT4G14640 CAM8; CAM8 (CALMODULIN 8); calcium ion binding;
K13448 calcium-binding protein CML
Length=151
Score = 179 bits (453), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 83/120 (69%), Positives = 104/120 (86%), Gaps = 0/120 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
IT +EL TV+RSL QNPTE EL D+I E+D+D NGTI+F EFL LMA+K++++D+E EL
Sbjct 29 ITVEELATVIRSLDQNPTEQELHDIITEIDSDSNGTIEFAEFLNLMAKKLQESDAEEELK 88
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
EAFKVFD+D NG ISA+EL HVM NLGEKLTDEEV++MI+EAD+DGDGQ+NY+EFV MM+
Sbjct 89 EAFKVFDKDQNGYISASELSHVMINLGEKLTDEEVEQMIKEADLDGDGQVNYDEFVKMMI 148
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DG+G I+ EL V+ +L + T++E+ ++I E D D +G I + EF+
Sbjct 13 EFKEAFCLFDKDGDGCITVEELATVIRSLDQNPTEQELHDIITEIDSDSNGTIEFAEFLN 72
Query 174 MMLAK 178
+M K
Sbjct 73 LMAKK 77
> cel:C13C12.1 cal-1; CALmodulin related genes family member (cal-1)
Length=161
Score = 162 bits (410), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 77/120 (64%), Positives = 98/120 (81%), Gaps = 1/120 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
I+TKELG MRSLGQNPTE E+ +MINEVD DGNG I+FPEF +M R MK+TDSE +
Sbjct 40 TISTKELGIAMRSLGQNPTEQEILEMINEVDIDGNGQIEFPEFCVMMKRMMKETDSEM-I 98
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFD+DGNG+I+A E R+ M ++G + ++EEVDEMI+E D+DGDG+I+YEEFV MM
Sbjct 99 REAFRVFDKDGNGVITAQEFRYFMVHMGMQFSEEEVDEMIKEVDVDGDGEIDYEEFVKMM 158
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 34/62 (54%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DGNG IS EL M +LG+ T++E+ EMI E DIDG+GQI + EF
Sbjct 25 EFREAFMMFDKDGNGTISTKELGIAMRSLGQNPTEQEILEMINEVDIDGNGQIEFPEFCV 84
Query 174 MM 175
MM
Sbjct 85 MM 86
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 43 QRAARVLLRMFKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLM 102
+ A RV + IT +E M +G +E E+ +MI EVD DG+G ID+ EF+ +M
Sbjct 99 REAFRVFDKDGNGVITAQEFRYFMVHMGMQFSEEEVDEMIKEVDVDGDGEIDYEEFVKMM 158
Query 103 ARK 105
+ +
Sbjct 159 SNQ 161
> cel:Y73B3A.12 hypothetical protein
Length=116
Score = 159 bits (402), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 78/86 (90%), Positives = 82/86 (95%), Gaps = 0/86 (0%)
Query 81 MINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMT 140
MINEVDADGNGTIDFPEFLT+MARKMK TDSE E+ EAF+VFD+DGNG ISAAELRHVMT
Sbjct 1 MINEVDADGNGTIDFPEFLTVMARKMKGTDSEEEIREAFRVFDKDGNGFISAAELRHVMT 60
Query 141 NLGEKLTDEEVDEMIREADIDGDGQI 166
NLGEKLTDEEVDEMIREADIDGDGQI
Sbjct 61 NLGEKLTDEEVDEMIREADIDGDGQI 86
Score = 34.7 bits (78), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 35 GPLCAQRPQRAARVLLRMFKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTI 93
G + + A RV + I+ EL VM +LG+ T+ E+ +MI E D DG+G I
Sbjct 28 GTDSEEEIREAFRVFDKDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQI 86
> sce:YBR109C CMD1; Calmodulin; Ca++ binding protein that regulates
Ca++ independent processes (mitosis, bud growth, actin
organization, endocytosis, etc.) and Ca++ dependent processes
(stress-activated pathways), targets include Nuf1p, Myo2p
and calcineurin; K02183 calmodulin
Length=147
Score = 157 bits (397), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 72/120 (60%), Positives = 100/120 (83%), Gaps = 1/120 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
+I++ EL TVMRSLG +P+EAE+ D++NE+D DGN I+F EFL LM+R++K DSE EL
Sbjct 27 SISSSELATVMRSLGLSPSEAEVNDLMNEIDVDGNHQIEFSEFLALMSRQLKSNDSEQEL 86
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
+EAFKVFD++G+GLISAAEL+HV+T++GEKLTD EVD+M+RE DG G+IN ++F ++
Sbjct 87 LEAFKVFDKNGDGLISAAELKHVLTSIGEKLTDAEVDDMLREVS-DGSGEINIQQFAALL 145
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 44/63 (69%), Gaps = 0/63 (0%)
Query 113 GELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
E EAF +FD+D NG IS++EL VM +LG ++ EV++++ E D+DG+ QI + EF+
Sbjct 11 AEFKEAFALFDKDNNGSISSSELATVMRSLGLSPSEAEVNDLMNEIDVDGNHQIEFSEFL 70
Query 173 GMM 175
+M
Sbjct 71 ALM 73
> cel:C18E9.1 cal-2; CALmodulin related genes family member (cal-2);
K02183 calmodulin
Length=171
Score = 154 bits (390), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 76/122 (62%), Positives = 95/122 (77%), Gaps = 1/122 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
+I++KELG MRSLGQNPTE EL DM+NEVD DG+GTIDF EF +M R K+ DSE +
Sbjct 51 SISSKELGVAMRSLGQNPTEQELLDMVNEVDIDGSGTIDFGEFCQMMKRMNKENDSE-MI 109
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
EAF+VFDRDGNG I+A E R+ MT++G++ +D+EVDE+I E DIDGDGQI+YEEF
Sbjct 110 REAFRVFDRDGNGFITADEFRYFMTHMGDQFSDQEVDEIIAEIDIDGDGQIDYEEFASTF 169
Query 176 LA 177
A
Sbjct 170 SA 171
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/62 (48%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E AF++FD+DGNG IS+ EL M +LG+ T++E+ +M+ E DIDG G I++ EF
Sbjct 36 EYKAAFRLFDKDGNGSISSKELGVAMRSLGQNPTEQELLDMVNEVDIDGSGTIDFGEFCQ 95
Query 174 MM 175
MM
Sbjct 96 MM 97
Score = 38.5 bits (88), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 43 QRAARVLLRMFKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLM 102
+ A RV R IT E M +G ++ E+ ++I E+D DG+G ID+ EF +
Sbjct 110 REAFRVFDRDGNGFITADEFRYFMTHMGDQFSDQEVDEIIAEIDIDGDGQIDYEEFASTF 169
Query 103 A 103
+
Sbjct 170 S 170
> ath:AT2G41090 calmodulin-like calcium-binding protein, 22 kDa
(CaBP-22); K13448 calcium-binding protein CML
Length=191
Score = 142 bits (358), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 75/122 (61%), Positives = 93/122 (76%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
ITT+E G VMRSLG N T+AELQ+ IN+ D DG+GTI+F EFL MA KDT SE +L
Sbjct 28 ITTEEFGAVMRSLGLNLTQAELQEEINDSDLDGDGTINFTEFLCAMA---KDTYSEKDLK 84
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
+ F++FD D NG ISAAE+R+V T L K TDEE+DE+I+ AD+DGDGQINY EF +M+
Sbjct 85 KDFRLFDIDKNGFISAAEMRYVRTILRWKQTDEEIDEIIKAADVDGDGQINYREFARLMM 144
Query 177 AK 178
AK
Sbjct 145 AK 146
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E E F V+D++G+G I+ E VM +LG LT E+ E I ++D+DGDG IN+ EF+
Sbjct 12 EFREQFSVYDKNGDGHITTEEFGAVMRSLGLNLTQAELQEEINDSDLDGDGTINFTEFLC 71
Query 174 MM 175
M
Sbjct 72 AM 73
Score = 33.9 bits (76), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSE 112
I+ E+ V L T+ E+ ++I D DG+G I++ EF LM K + D++
Sbjct 98 ISAAEMRYVRTILRWKQTDEEIDEIIKAADVDGDGQINYREFARLMMAKNQGHDTK 153
> cel:T07G12.1 cal-4; CALmodulin related genes family member (cal-4);
K02183 calmodulin
Length=182
Score = 136 bits (342), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 74/143 (51%), Positives = 94/143 (65%), Gaps = 3/143 (2%)
Query 34 FGPLCAQRPQRAARVLLRMFKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTI 93
F P Q +A ++ + + KELG MR LG NPTE EL +M+NE D DGNG I
Sbjct 17 FTPEELQEFAQAFKLFDKDGNNTMNIKELGEAMRMLGLNPTEEELLNMVNEYDVDGNGKI 76
Query 94 DFPEFLTLMARKMKDTDSEGELIE-AFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVD 152
DF EF +M K+TD E LI AFKVFD+DGNG I+A E +H MT +GE+ ++EEVD
Sbjct 77 DFGEFCKMMKEMNKETDQE--LIRLAFKVFDKDGNGYITAQEFKHFMTTMGERFSEEEVD 134
Query 153 EMIREADIDGDGQINYEEFVGMM 175
E+IRE D DGD QI+ +EFV M+
Sbjct 135 EIIREVDKDGDEQIDLDEFVNMV 157
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 11/86 (12%)
Query 34 FGPLCA-------QRPQRAARVLLRMFKL----AITTKELGTVMRSLGQNPTEAELQDMI 82
FG C + Q R+ ++F IT +E M ++G+ +E E+ ++I
Sbjct 78 FGEFCKMMKEMNKETDQELIRLAFKVFDKDGNGYITAQEFKHFMTTMGERFSEEEVDEII 137
Query 83 NEVDADGNGTIDFPEFLTLMARKMKD 108
EVD DG+ ID EF+ ++A + D
Sbjct 138 REVDKDGDEQIDLDEFVNMVAPIVSD 163
> ath:AT2G41100 TCH3; TCH3 (TOUCH 3); calcium ion binding; K13448
calcium-binding protein CML
Length=289
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 74/140 (52%), Positives = 91/140 (65%), Gaps = 17/140 (12%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMA---------RKM 106
+IT KEL TVM SLG+N T+A+LQDM+NEVD DG+GTIDFPEFL LMA R
Sbjct 81 SITKKELRTVMFSLGKNRTKADLQDMMNEVDLDGDGTIDFPEFLYLMAKNQGHDQAPRHT 140
Query 107 KDT--------DSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREA 158
K T D E EAF+VFD++G+G I+ ELR M +LGE T E+ +MI EA
Sbjct 141 KKTMVDYQLTDDQILEFREAFRVFDKNGDGYITVNELRTTMRSLGETQTKAELQDMINEA 200
Query 159 DIDGDGQINYEEFVGMMLAK 178
D DGDG I++ EFV +M K
Sbjct 201 DADGDGTISFSEFVCVMTGK 220
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 84/127 (66%), Gaps = 16/127 (12%)
Query 65 VMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMA---------RKMKDT------ 109
+MRS+G+ PT+A+LQD++NE D DG+GTIDFPEFL +MA R K T
Sbjct 1 MMRSIGEKPTKADLQDLMNEADLDGDGTIDFPEFLCVMAKNQGHDQAPRHTKKTMADKLT 60
Query 110 -DSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINY 168
D E E+F++FD++G+G I+ ELR VM +LG+ T ++ +M+ E D+DGDG I++
Sbjct 61 DDQITEYRESFRLFDKNGDGSITKKELRTVMFSLGKNRTKADLQDMMNEVDLDGDGTIDF 120
Query 169 EEFVGMM 175
EF+ +M
Sbjct 121 PEFLYLM 127
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/70 (57%), Positives = 47/70 (67%), Gaps = 0/70 (0%)
Query 45 AARVLLRMFKLAITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMAR 104
A RV + IT EL T MRSLG+ T+AELQDMINE DADG+GTI F EF+ +M
Sbjct 160 AFRVFDKNGDGYITVNELRTTMRSLGETQTKAELQDMINEADADGDGTISFSEFVCVMTG 219
Query 105 KMKDTDSEGE 114
KM DT S+ E
Sbjct 220 KMIDTQSKKE 229
> xla:379095 tnnc2, MGC53213; troponin C type 2 (fast); K05865
troponin C, slow skeletal and cardiac muscles
Length=163
Score = 126 bits (316), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 62/122 (50%), Positives = 90/122 (73%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDT---DSEG 113
I+TKELGTVMR LGQ PT+ EL +I EVD DG+GTIDF EFL +M R+MK+ SE
Sbjct 38 ISTKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLVMMVRQMKEDAQGKSEE 97
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
EL E F++FD++ +G I EL ++ + GE +TDEE++E++++ D + DG+I+++EF+
Sbjct 98 ELAECFRIFDKNADGYIDGEELAEILRSSGESITDEEIEELMKDGDKNNDGKIDFDEFLK 157
Query 174 MM 175
MM
Sbjct 158 MM 159
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E AF +FD DG G IS EL VM LG+ T EE+D +I E D DG G I++EEF+
Sbjct 22 EFKAAFDMFDTDGGGDISTKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLV 81
Query 174 MML 176
MM+
Sbjct 82 MMV 84
> xla:379085 tnnc2, MGC52923; fast skeletal troponin C beta; K12042
troponin C, skeletal muscle
Length=163
Score = 126 bits (316), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 62/122 (50%), Positives = 90/122 (73%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDT---DSEG 113
I+TKELGTVMR LGQ PT+ EL +I EVD DG+GTIDF EFL +M R+MK+ SE
Sbjct 38 ISTKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLVMMVRQMKEDAQGKSEE 97
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
EL E F++FD++ +G I EL ++ + GE +TDEE++E++++ D + DG+I+++EF+
Sbjct 98 ELAECFRIFDKNADGYIDGEELAEILRSSGESITDEEIEELMKDGDKNNDGKIDFDEFLK 157
Query 174 MM 175
MM
Sbjct 158 MM 159
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E AF +FD DG G IS EL VM LG+ T EE+D +I E D DG G I++EEF+
Sbjct 22 EFKAAFDMFDTDGGGDISTKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLV 81
Query 174 MML 176
MM+
Sbjct 82 MMV 84
> cel:M02B7.6 cal-3; CALmodulin related genes family member (cal-3)
Length=234
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 65/117 (55%), Positives = 89/117 (76%), Gaps = 1/117 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
I+ KELG MR+LGQNPTE ++ ++I++VD DGNG ++FPEF +M R MK+TDSE +
Sbjct 115 TISIKELGVAMRALGQNPTEQQMMEIIHDVDLDGNGQVEFPEFCVMMKRIMKETDSEM-I 173
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
EAFK+FDRDGNG+I+A E + M N+G + EV+EM+ E D DG+G+I+YEEFV
Sbjct 174 REAFKIFDRDGNGVITANEFKLFMINMGMCFDEVEVEEMMNEVDCDGNGEIDYEEFV 230
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 42/62 (67%), Gaps = 0/62 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF +FD+DGNG IS EL M LG+ T++++ E+I + D+DG+GQ+ + EF
Sbjct 100 EFKEAFLLFDKDGNGTISIKELGVAMRALGQNPTEQQMMEIIHDVDLDGNGQVEFPEFCV 159
Query 174 MM 175
MM
Sbjct 160 MM 161
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 47/95 (49%), Gaps = 9/95 (9%)
Query 86 DADGNGTIDFPEFLTLMARKMKDTDSEGELIEAFKVFDRDGNGLISAAELRHVMTNLGEK 145
D DGNGTI E L + R + +E +++E D DGNG + E +M ++
Sbjct 109 DKDGNGTISIKE-LGVAMRALGQNPTEQQMMEIIHDVDLDGNGQVEFPEFCVMM----KR 163
Query 146 LTDEEVDEMIREA----DIDGDGQINYEEFVGMML 176
+ E EMIREA D DG+G I EF M+
Sbjct 164 IMKETDSEMIREAFKIFDRDGNGVITANEFKLFMI 198
> xla:399104 tnnc1, MGC80066; cardiac troponin C; K05865 troponin
C, slow skeletal and cardiac muscles
Length=161
Score = 122 bits (305), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 61/122 (50%), Positives = 90/122 (73%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTD---SEG 113
I+TKELG VMR LGQNPT ELQ+MI+EVD DG+GT+DF EFL +M R MKD SE
Sbjct 36 ISTKELGKVMRMLGQNPTPEELQEMIDEVDEDGSGTVDFDEFLVMMVRCMKDDSKGKSEE 95
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
EL + F++FD++ +G I EL+ ++ GE +T+++++E++R+ D + DG+I+Y+EF+
Sbjct 96 ELSDLFRMFDKNADGYIDLDELKMMLEATGETITEDDIEELMRDGDKNNDGRIDYDEFLE 155
Query 174 MM 175
M
Sbjct 156 FM 157
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 118 AFKVFDRDG-NGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
AF +F +D +G IS EL VM LG+ T EE+ EMI E D DG G ++++EF+ MM+
Sbjct 23 AFDIFVQDAEDGCISTKELGKVMRMLGQNPTPEELQEMIDEVDEDGSGTVDFDEFLVMMV 82
> mmu:21925 Tnnc2, Tncs; troponin C2, fast; K12042 troponin C,
skeletal muscle
Length=160
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 61/122 (50%), Positives = 87/122 (71%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDT---DSEG 113
I+ KELGTVMR LGQ PT+ EL +I EVD DG+GTIDF EFL +M R+MK+ SE
Sbjct 35 ISVKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLVMMVRQMKEDAKGKSEE 94
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
EL E F++FDR+ +G I A EL + GE +T+EE++ ++++ D + DG+I+++EF+
Sbjct 95 ELAECFRIFDRNADGYIDAEELAEIFRASGEHVTEEEIESLMKDGDKNNDGRIDFDEFLK 154
Query 174 MM 175
MM
Sbjct 155 MM 156
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E AF +FD DG G IS EL VM LG+ T EE+D +I E D DG G I++EEF+
Sbjct 19 EFKAAFDMFDADGGGDISVKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLV 78
Query 174 MML 176
MM+
Sbjct 79 MMV 81
> ath:AT3G51920 CAM9; CAM9 (CALMODULIN 9); calcium ion binding;
K13448 calcium-binding protein CML
Length=151
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/121 (49%), Positives = 85/121 (70%), Gaps = 0/121 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGELI 116
IT ++L VM+S+G+NP +LQ M+++VD GNG I F +FL +MA+ + ELI
Sbjct 28 ITKEKLTKVMKSMGKNPKAEQLQQMMSDVDIFGNGGITFDDFLYIMAQNTSQESASDELI 87
Query 117 EAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
E F+VFDRDG+GLIS EL M ++G K+T EE + M+READ+DGDG +++ EF MM+
Sbjct 88 EVFRVFDRDGDGLISQLELGEGMKDMGMKITAEEAEHMVREADLDGDGFLSFHEFSKMMI 147
Query 177 A 177
A
Sbjct 148 A 148
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E EAF + D+D +G I+ +L VM ++G+ E++ +M+ + DI G+G I +++F+
Sbjct 12 EFYEAFCLIDKDSDGFITKEKLTKVMKSMGKNPKAEQLQQMMSDVDIFGNGGITFDDFLY 71
Query 174 MM 175
+M
Sbjct 72 IM 73
> dre:353247 tnnc1a, cTnC, tnnc1, zgc:103465; troponin C type
1a (slow); K05865 troponin C, slow skeletal and cardiac muscles
Length=161
Score = 121 bits (303), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 62/123 (50%), Positives = 91/123 (73%), Gaps = 5/123 (4%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEG--- 113
I+TKELG VMR LGQNPT ELQ+MI+EVD DG+G +DF EFL +M R MKD DS+G
Sbjct 36 ISTKELGKVMRMLGQNPTPEELQEMIDEVDEDGSGAVDFEEFLVMMVRCMKD-DSKGRPE 94
Query 114 -ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFV 172
EL E F++FD++ +G I EL+ ++ GE +T+++++E++R+ D + DG+I+Y+EF+
Sbjct 95 EELAELFRMFDKNADGYIDLDELKLMLEATGEAITEDDIEELMRDGDKNNDGKIDYDEFL 154
Query 173 GMM 175
M
Sbjct 155 EFM 157
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 118 AFKVFDRDG-NGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMML 176
AF +F +D +G IS EL VM LG+ T EE+ EMI E D DG G +++EEF+ MM+
Sbjct 23 AFDIFVQDAEDGCISTKELGKVMRMLGQNPTPEELQEMIDEVDEDGSGAVDFEEFLVMMV 82
> xla:735211 cetn4, MGC130946; centrin 4
Length=171
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 60/120 (50%), Positives = 85/120 (70%), Gaps = 0/120 (0%)
Query 56 AITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTDSEGEL 115
I KEL MR+LG P + E++ +I+++D DG+G IDF +FL+LM +KM + DS+ E+
Sbjct 46 TIDVKELKVAMRALGFEPKKEEMKKIISDIDKDGSGIIDFEDFLSLMTQKMSEKDSKEEI 105
Query 116 IEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVGMM 175
++AF++FD D G IS L+ V LGE LTDEE+ EMI EAD DGDG+IN +EF+ +M
Sbjct 106 MKAFRLFDDDNTGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEINEQEFLRIM 165
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E+ EAF +FD DG G I EL+ M LG + EE+ ++I + D DG G I++E+F+
Sbjct 31 EIREAFDLFDTDGTGTIDVKELKVAMRALGFEPKKEEMKKIISDIDKDGSGIIDFEDFLS 90
Query 174 MMLAK 178
+M K
Sbjct 91 LMTQK 95
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMAR 104
I+ K L V + LG+N T+ ELQ+MI+E D DG+G I+ EFL +M +
Sbjct 120 ISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEINEQEFLRIMRK 167
> hsa:7125 TNNC2; troponin C type 2 (fast); K12042 troponin C,
skeletal muscle
Length=160
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 61/122 (50%), Positives = 86/122 (70%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDT---DSEG 113
I+ KELGTVMR LGQ PT+ EL +I EVD DG+GTIDF EFL +M R+MK+ SE
Sbjct 35 ISVKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLVMMVRQMKEDAKGKSEE 94
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
EL E F++FDR+ +G I EL + GE +TDEE++ ++++ D + DG+I+++EF+
Sbjct 95 ELAECFRIFDRNADGYIDPEELAEIFRASGEHVTDEEIESLMKDGDKNNDGRIDFDEFLK 154
Query 174 MM 175
MM
Sbjct 155 MM 156
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
E AF +FD DG G IS EL VM LG+ T EE+D +I E D DG G I++EEF+
Sbjct 19 EFKAAFDMFDADGGGDISVKELGTVMRMLGQTPTKEELDAIIEEVDEDGSGTIDFEEFLV 78
Query 174 MML 176
MM+
Sbjct 79 MMV 81
> mmu:21924 Tnnc1, AI874626, TnC, cTnC, cTnI, tncc; troponin C,
cardiac/slow skeletal; K05865 troponin C, slow skeletal and
cardiac muscles
Length=161
Score = 120 bits (301), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 60/122 (49%), Positives = 90/122 (73%), Gaps = 3/122 (2%)
Query 57 ITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTLMARKMKDTD---SEG 113
I+TKELG VMR LGQNPT ELQ+MI+EVD DG+GT+DF EFL +M R MKD SE
Sbjct 36 ISTKELGKVMRMLGQNPTPEELQEMIDEVDEDGSGTVDFDEFLVMMVRCMKDDSKGKSEE 95
Query 114 ELIEAFKVFDRDGNGLISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQINYEEFVG 173
EL + F++FD++ +G I EL+ ++ GE +T+++++E++++ D + DG+I+Y+EF+
Sbjct 96 ELSDLFRMFDKNADGYIDLDELKMMLQATGETITEDDIEELMKDGDKNNDGRIDYDEFLE 155
Query 174 MM 175
M
Sbjct 156 FM 157
Lambda K H
0.320 0.137 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4730349484
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40