bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1002_orf2
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
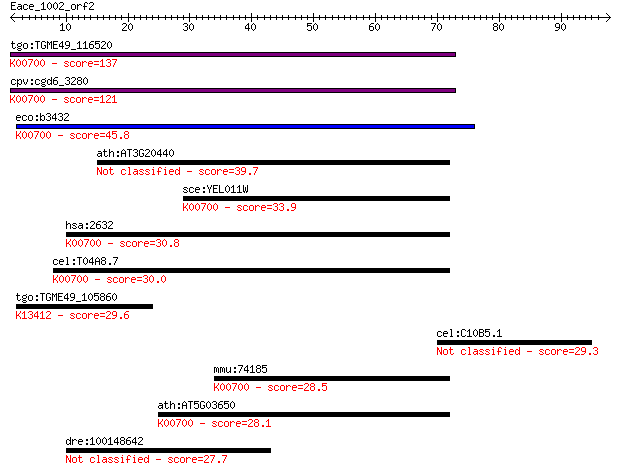
tgo:TGME49_116520 1,4-alpha-glucan branching enzyme, putative ... 137 1e-32
cpv:cgd6_3280 glycogen branching enzyme (1,4-alpha-glucan bran... 121 7e-28
eco:b3432 glgB, ECK3418, JW3395; 1,4-alpha-glucan branching en... 45.8 3e-05
ath:AT3G20440 EMB2729; alpha-amylase/ catalytic/ cation binding 39.7 0.003
sce:YEL011W GLC3, GHA1; Glc3p (EC:2.4.1.18); K00700 1,4-alpha-... 33.9 0.12
hsa:2632 GBE1, GBE; glucan (1,4-alpha-), branching enzyme 1 (E... 30.8 1.0
cel:T04A8.7 hypothetical protein; K00700 1,4-alpha-glucan bran... 30.0 1.7
tgo:TGME49_105860 calcium-dependent protein kinase 1, putative... 29.6 2.5
cel:C10B5.1 hypothetical protein 29.3 3.1
mmu:74185 Gbe1, 2310045H19Rik, 2810426P10Rik, D16Ertd536e; glu... 28.5 5.0
ath:AT5G03650 SBE2.2; SBE2.2 (starch branching enzyme 2.2); 1,... 28.1 7.2
dre:100148642 phosphatidylinositol glycan, class V-like 27.7 8.6
> tgo:TGME49_116520 1,4-alpha-glucan branching enzyme, putative
(EC:2.4.1.18); K00700 1,4-alpha-glucan branching enzyme [EC:2.4.1.18]
Length=972
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 57/72 (79%), Positives = 66/72 (91%), Gaps = 0/72 (0%)
Query 1 GDDETWNFKWTDCDNNNDCIVAFLRSYEFWFNDILVVCNFSPQAYYRYPVGVPHGGEWQL 60
GDDE WNFKWTDCDN+ DC++AFLRSY W+NDILVVCNFSP YYRYP+GVPHGGEW++
Sbjct 802 GDDEPWNFKWTDCDNSKDCVIAFLRSYMDWYNDILVVCNFSPNVYYRYPIGVPHGGEWEV 861
Query 61 LLNSDDWKYAGG 72
+LNSDDW+YAGG
Sbjct 862 ILNSDDWRYAGG 873
> cpv:cgd6_3280 glycogen branching enzyme (1,4-alpha-glucan branching
enzyme) ; K00700 1,4-alpha-glucan branching enzyme [EC:2.4.1.18]
Length=1030
Score = 121 bits (303), Expect = 7e-28, Method: Composition-based stats.
Identities = 47/72 (65%), Positives = 60/72 (83%), Gaps = 0/72 (0%)
Query 1 GDDETWNFKWTDCDNNNDCIVAFLRSYEFWFNDILVVCNFSPQAYYRYPVGVPHGGEWQL 60
GDDE+WNF+W DC+N+ DCI+AFLR Y+ W+ND++VVCNFS + Y YP+GVPHG EW +
Sbjct 912 GDDESWNFQWVDCENSQDCIIAFLRKYKEWYNDVVVVCNFSSRRYNHYPIGVPHGKEWLV 971
Query 61 LLNSDDWKYAGG 72
+LNSDDWKY G
Sbjct 972 MLNSDDWKYGGA 983
> eco:b3432 glgB, ECK3418, JW3395; 1,4-alpha-glucan branching
enzyme (EC:2.4.1.18); K00700 1,4-alpha-glucan branching enzyme
[EC:2.4.1.18]
Length=728
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Query 2 DDETWNFKWTDCDNNNDCIVAFLRSYEFWFNDILVVCNFSPQAYYRYPVGVPHGGEWQLL 61
D + + F+W D+ ++ F+R + N+I+V NF+P + Y G+ G+W+ +
Sbjct 620 DFDPYGFEWLVVDDKERSVLIFVRR-DKEGNEIIVASNFTPVPRHDYRFGINQPGKWREI 678
Query 62 LNSDDWKYAGGCAA 75
LN+D Y G A
Sbjct 679 LNTDSMHYHGSNAG 692
> ath:AT3G20440 EMB2729; alpha-amylase/ catalytic/ cation binding
Length=899
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 7/58 (12%)
Query 15 NNNDCIVAFLRSYEFWFNDILVVCNFSP-QAYYRYPVGVPHGGEWQLLLNSDDWKYAG 71
N+ + +++F R L + NF P +Y +Y VGV GE+ ++LNSD+ KY G
Sbjct 803 NDANMVISFSRG------PFLFIFNFHPSNSYEKYDVGVEEAGEYTMILNSDEVKYGG 854
> sce:YEL011W GLC3, GHA1; Glc3p (EC:2.4.1.18); K00700 1,4-alpha-glucan
branching enzyme [EC:2.4.1.18]
Length=704
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query 29 FWFNDILVVCNFSP-QAYYRYPVGVPHGGEWQLLLNSDDWKYAG 71
F N++L + NF P +Y Y VGV G + ++LNSD ++ G
Sbjct 621 FERNNLLFIFNFHPTNSYSDYRVGVEKAGTYHIVLNSDRAEFGG 664
> hsa:2632 GBE1, GBE; glucan (1,4-alpha-), branching enzyme 1
(EC:2.4.1.18); K00700 1,4-alpha-glucan branching enzyme [EC:2.4.1.18]
Length=702
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 34/63 (53%), Gaps = 7/63 (11%)
Query 10 WTDCDNNNDCIVAFLRSYEFWFNDILVVCNFSP-QAYYRYPVGVPHGGEWQLLLNSDDWK 68
+ + + I+AF R+ +L + NF P ++Y Y VG G+++++L+SD +
Sbjct 603 YVSEKHEGNKIIAFERA------GLLFIFNFHPSKSYTDYRVGTALPGKFKIVLDSDAAE 656
Query 69 YAG 71
Y G
Sbjct 657 YGG 659
> cel:T04A8.7 hypothetical protein; K00700 1,4-alpha-glucan branching
enzyme [EC:2.4.1.18]
Length=681
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query 8 FKWTDCDNNNDCIVAFLRSYEFWFNDILVVCNFSP-QAYYRYPVGVPHGGEWQLLLNSDD 66
+ +T ++ D + F R ++ V N P +++ Y +GV G +++ LNSD+
Sbjct 582 YAYTSWKHDGDKTIVFERG------GLVFVINLHPTKSFADYSIGVNTPGRYRIALNSDE 635
Query 67 WKYAG 71
K+ G
Sbjct 636 SKFGG 640
> tgo:TGME49_105860 calcium-dependent protein kinase 1, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=537
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 12/22 (54%), Positives = 14/22 (63%), Gaps = 0/22 (0%)
Query 2 DDETWNFKWTDCDNNNDCIVAF 23
DDETW+ +CD NND V F
Sbjct 499 DDETWHQVLQECDKNNDGEVDF 520
> cel:C10B5.1 hypothetical protein
Length=311
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 70 AGGCAAWAMAPISTQLRAAGWDGPI 94
A GC+A + +P S Q+ A WDG I
Sbjct 236 AAGCSAISFSPNSKQIIAGFWDGSI 260
> mmu:74185 Gbe1, 2310045H19Rik, 2810426P10Rik, D16Ertd536e; glucan
(1,4-alpha-), branching enzyme 1 (EC:2.4.1.18); K00700
1,4-alpha-glucan branching enzyme [EC:2.4.1.18]
Length=702
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 34 ILVVCNFSP-QAYYRYPVGVPHGGEWQLLLNSDDWKYAG 71
+L + NF P ++Y Y VG G+++++L+SD +Y G
Sbjct 621 LLFIFNFHPSKSYTDYRVGTATPGKFKIVLDSDAAEYGG 659
> ath:AT5G03650 SBE2.2; SBE2.2 (starch branching enzyme 2.2);
1,4-alpha-glucan branching enzyme (EC:2.4.1.18); K00700 1,4-alpha-glucan
branching enzyme [EC:2.4.1.18]
Length=805
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query 25 RSYEFWFNDILVVCNFS-PQAYYRYPVGVPHGGEWQLLLNSDDWKYAG 71
R F D++ V NF +Y+ Y +G G+++++L+SDD + G
Sbjct 716 RVIVFERGDLVFVFNFHWTSSYFDYRIGCSKPGKYKIVLDSDDPLFGG 763
> dre:100148642 phosphatidylinositol glycan, class V-like
Length=523
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 16/33 (48%), Gaps = 2/33 (6%)
Query 10 WTDCDNNNDCIVAFLRSYEFWFNDILVVCNFSP 42
W C CI+ + SY WF + + CNF P
Sbjct 491 WKTCSIYTQCILGYFISY--WFLGLALHCNFLP 521
Lambda K H
0.322 0.140 0.520
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40