bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1067_orf2
Length=212
Score E
Sequences producing significant alignments: (Bits) Value
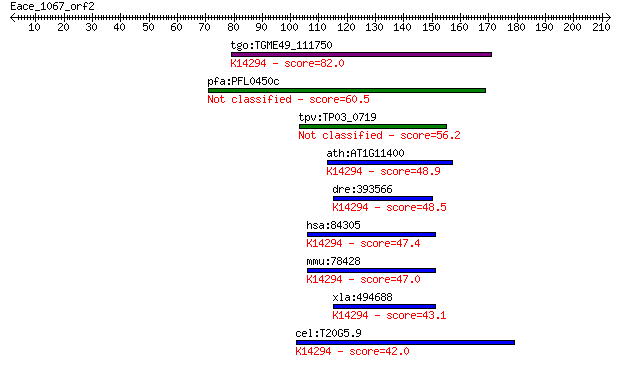
tgo:TGME49_111750 hypothetical protein ; K14294 partner of Y14... 82.0 1e-15
pfa:PFL0450c conserved Plasmodium protein 60.5 4e-09
tpv:TP03_0719 hypothetical protein 56.2 9e-08
ath:AT1G11400 PYM; PYM (Partner of Y14-Mago); protein binding;... 48.9 1e-05
dre:393566 MGC65891, wibg, zgc:77281; zgc:65891; K14294 partne... 48.5 1e-05
hsa:84305 WIBG, MGC13064, PYM; within bgcn homolog (Drosophila... 47.4 4e-05
mmu:78428 Wibg, A030010B05Rik, Pym; within bgcn homolog (Droso... 47.0 5e-05
xla:494688 wibg; within bgcn homolog; K14294 partner of Y14 an... 43.1 7e-04
cel:T20G5.9 hypothetical protein; K14294 partner of Y14 and mago 42.0 0.002
> tgo:TGME49_111750 hypothetical protein ; K14294 partner of Y14
and mago
Length=250
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 46/92 (50%), Positives = 58/92 (63%), Gaps = 4/92 (4%)
Query 79 FGELLCSSSSNGSSDVLYSQQTAGGETYLLDPTTGNKYIPASVRPDGSLRKPIRVRAGYT 138
F EL S S+GS V ++T GE YL+D TTG K IP S RPDG+ RK IRVRAGY
Sbjct 21 FPEL--DSRSDGSLRV--EERTDMGERYLVDMTTGEKIIPGSRRPDGTYRKEIRVRAGYV 76
Query 139 PREEMSGYRTRHQLSQEQHRKTVGSSIPGLAP 170
P EE ++TR QLS+ + + +IPG +P
Sbjct 77 PLEERRTFQTRQQLSRNERQVPGAGNIPGFSP 108
> pfa:PFL0450c conserved Plasmodium protein
Length=167
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 5/103 (4%)
Query 71 MQTPPPSGFG-----ELLCSSSSNGSSDVLYSQQTAGGETYLLDPTTGNKYIPASVRPDG 125
MQ P +GF L + N + T G+ Y+++ T K+I + R DG
Sbjct 1 MQKKPSTGFKLENTYRALFDNDDNNEKSYIREVVTPLGDVYIINEKTQEKFIKGTQRSDG 60
Query 126 SLRKPIRVRAGYTPREEMSGYRTRHQLSQEQHRKTVGSSIPGL 168
+ RK IRV+ Y P+EE Y+ + +L +EQ+R ++ P +
Sbjct 61 TFRKNIRVKTDYMPQEENCAYQVKGKLLEEQNRLLTKNTSPNV 103
> tpv:TP03_0719 hypothetical protein
Length=393
Score = 56.2 bits (134), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 26/52 (50%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 103 GETYLLDPTTGNKYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTRHQLSQ 154
GE+Y D +TG KYI S RPDG+ RK I++R GY P EE Y H+ +Q
Sbjct 26 GESYFTDESTGGKYIIPSKRPDGTYRKEIKIRPGYVPPEERQLYVPLHRRTQ 77
> ath:AT1G11400 PYM; PYM (Partner of Y14-Mago); protein binding;
K14294 partner of Y14 and mago
Length=204
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 32/44 (72%), Gaps = 0/44 (0%)
Query 113 GNKYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTRHQLSQEQ 156
G + + + RPDG+LRKPIR+R GYTP +E+ Y+++ L +++
Sbjct 22 GERILEPTRRPDGTLRKPIRIRPGYTPEDEVVKYQSKGSLMKKE 65
> dre:393566 MGC65891, wibg, zgc:77281; zgc:65891; K14294 partner
of Y14 and mago
Length=194
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 115 KYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTR 149
KYI A+ RPDGS RKP RVR GY P+EE+ Y +
Sbjct 12 KYIAATQRPDGSWRKPRRVRDGYVPQEEVPVYENK 46
> hsa:84305 WIBG, MGC13064, PYM; within bgcn homolog (Drosophila);
K14294 partner of Y14 and mago
Length=203
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 21/45 (46%), Positives = 30/45 (66%), Gaps = 1/45 (2%)
Query 106 YLLDPTTGNKYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTRH 150
Y+ D TG KYI ++ RPDG+ RK RV+ GY P+EE+ Y ++
Sbjct 5 YVTD-ETGGKYIASTQRPDGTWRKQRRVKEGYVPQEEVPVYENKY 48
> mmu:78428 Wibg, A030010B05Rik, Pym; within bgcn homolog (Drosophila);
K14294 partner of Y14 and mago
Length=202
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/45 (46%), Positives = 30/45 (66%), Gaps = 1/45 (2%)
Query 106 YLLDPTTGNKYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTRH 150
Y+ D TG KYI ++ RPDG+ RK RV+ GY P+EE+ Y ++
Sbjct 5 YVTD-ETGGKYIASTQRPDGTWRKQRRVKEGYVPQEEVPVYENKY 48
> xla:494688 wibg; within bgcn homolog; K14294 partner of Y14
and mago
Length=199
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 115 KYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTRH 150
KYI ++ RPDGS RK +V+ GY P+EE+ Y ++
Sbjct 12 KYIASTQRPDGSWRKQRKVKEGYVPQEEVPVYENKY 47
> cel:T20G5.9 hypothetical protein; K14294 partner of Y14 and
mago
Length=164
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 42/78 (53%), Gaps = 6/78 (7%)
Query 102 GGETYLLDPTTGNKYIPASVRPDGSLRKPIRVRAGYTPREEMSGYRTRHQLSQEQHRKTV 161
GG+ ++ G +I A+ R DG+ RK RV+ GY P++E Y+ + QL R
Sbjct 17 GGD-LRVETENGETFITATQRADGTWRKARRVKGGYIPQDEQPKYQNKMQLEATNGR--- 72
Query 162 GSSIP-GLAPLEDSSSSS 178
SS+P G+ P +S SS
Sbjct 73 -SSVPAGVNPRAMASGSS 89
Lambda K H
0.304 0.117 0.309
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6687805280
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40