bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1098_orf2
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
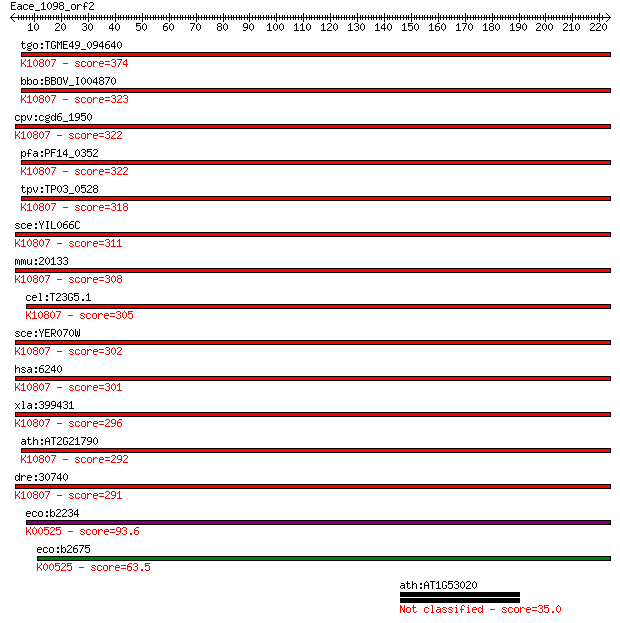
tgo:TGME49_094640 ribonucleoside-diphosphate reductase, large ... 374 1e-103
bbo:BBOV_I004870 19.m02176; ribonucleoside-diphosphate reducta... 323 4e-88
cpv:cgd6_1950 ribonucleotide-diphosphate reductase large chain... 322 5e-88
pfa:PF14_0352 ribonucleoside-diphosphate reductase, large subu... 322 6e-88
tpv:TP03_0528 ribonucleotide-diphosphate reductase large subun... 318 1e-86
sce:YIL066C RNR3, DIN1, RIR3; One of two large regulatory subu... 311 2e-84
mmu:20133 Rrm1, RnrM1; ribonucleotide reductase M1 (EC:1.17.4.... 308 7e-84
cel:T23G5.1 rnr-1; RiboNucleotide Reductase family member (rnr... 305 1e-82
sce:YER070W RNR1, CRT7, RIR1, SDS12; One of two large regulato... 302 6e-82
hsa:6240 RRM1, R1, RIR1, RR1; ribonucleotide reductase M1 (EC:... 301 1e-81
xla:399431 rrm1; ribonucleotide reductase M1 (EC:1.17.4.1); K1... 296 3e-80
ath:AT2G21790 RNR1; RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ATP bin... 292 7e-79
dre:30740 rrm1, CHUNP6866, cb396, cb838, r1, sb:cb548, wu:fb39... 291 2e-78
eco:b2234 nrdA, dnaF, ECK2226, JW2228; ribonucleoside-diphosph... 93.6 4e-19
eco:b2675 nrdE, ECK2669, JW2650; ribonucleoside-diphosphate re... 63.5 6e-10
ath:AT1G53020 UBC26; UBC26 (UBIQUITIN-CONJUGATING ENZYME 26); ... 35.0 0.23
> tgo:TGME49_094640 ribonucleoside-diphosphate reductase, large
subunit, putative (EC:5.1.3.4 1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=877
Score = 374 bits (961), Expect = 1e-103, Method: Compositional matrix adjust.
Identities = 169/219 (77%), Positives = 195/219 (89%), Gaps = 0/219 (0%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LWFAILQAQIETGTPYMLYKDACNRKSN ++LGT++ SNLC EVV+YTS +EVAVCNL
Sbjct 444 QHLWFAILQAQIETGTPYMLYKDACNRKSNQKNLGTIKCSNLCTEVVEYTSKDEVAVCNL 503
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGV 124
ASVSLP+FVD E +TFDY+ LK ++KVMTRNLN +IDRNYYP+PEA++SNLRHRP+GLGV
Sbjct 504 ASVSLPKFVDRESRTFDYEHLKRIVKVMTRNLNRVIDRNYYPVPEAKKSNLRHRPVGLGV 563
Query 125 QGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGV 184
QGLADAF+LLR PFDSP AR +N+NIFECIYFAA EAS ELAA G Y TYEGSP+S+G+
Sbjct 564 QGLADAFMLLRYPFDSPEARVLNRNIFECIYFAALEASCELAAEEGPYATYEGSPVSQGI 623
Query 185 FQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
QFDMWGV P SGLCDW GLR+K+K +G+RNSLLVSPMP
Sbjct 624 LQFDMWGVTPSSGLCDWDGLREKIKAHGVRNSLLVSPMP 662
> bbo:BBOV_I004870 19.m02176; ribonucleoside-diphosphate reductase
large chain (EC:1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=838
Score = 323 bits (827), Expect = 4e-88, Method: Compositional matrix adjust.
Identities = 142/219 (64%), Positives = 180/219 (82%), Gaps = 0/219 (0%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LWFAILQ+QIETG P+MLYKDACN KSN Q+LGT++SSNLCCE+VQYTSPEEVAVCNL
Sbjct 412 QTLWFAILQSQIETGNPFMLYKDACNAKSNQQNLGTIKSSNLCCEIVQYTSPEEVAVCNL 471
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGV 124
AS++LP +VD E KT+D+ KL DV +V+T NLN +IDRNYYP+PEA+ SN RHRPIG+GV
Sbjct 472 ASIALPMYVDKENKTYDFKKLYDVARVITYNLNKVIDRNYYPVPEAKVSNHRHRPIGVGV 531
Query 125 QGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGV 184
QGLAD F+L+R PF+S A+++N+ IFE IY+ + S+ LA +GTYE+Y GSP S+G
Sbjct 532 QGLADTFMLMRYPFESEEAKELNRRIFETIYYGCLDESISLAEKYGTYESYPGSPASQGK 591
Query 185 FQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
QFD+WG D+ L DW GL+Q++ K+GLRNSL ++PMP
Sbjct 592 LQFDLWGATVDNKLWDWDGLKQRMAKHGLRNSLFLAPMP 630
> cpv:cgd6_1950 ribonucleotide-diphosphate reductase large chain;
RIR1; c-terminal PFL-like glycyl radical enzymes-like fold
; K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=804
Score = 322 bits (826), Expect = 5e-88, Method: Compositional matrix adjust.
Identities = 142/221 (64%), Positives = 181/221 (81%), Gaps = 0/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LWF ILQAQIETGTP++ YKDA N KSN ++LGT+ SSNLC E+++YTS +EVAVC
Sbjct 384 KAQKLWFLILQAQIETGTPFICYKDAANSKSNQKNLGTIVSSNLCTEIIEYTSTDEVAVC 443
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS+ LP+FVD KTFD+DKLK+V KV+TRNLN +ID YY + E ++SNLRHRP+G+
Sbjct 444 NLASIGLPKFVDKNNKTFDFDKLKEVTKVITRNLNKLIDVGYYSLKECKKSNLRHRPLGI 503
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
G+QGLAD F++LR+P++S A+ +NK IFE IY+AA +AS ELA +G YETY GSP SK
Sbjct 504 GIQGLADCFMMLRMPYESEGAKKLNKQIFEVIYYAALDASCELAEKYGPYETYSGSPASK 563
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ QFDMWGV PDSGLCDW L+ ++ K+G+RNSLL+SPMP
Sbjct 564 GILQFDMWGVTPDSGLCDWDLLKDRISKHGIRNSLLISPMP 604
> pfa:PF14_0352 ribonucleoside-diphosphate reductase, large subunit;
K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=847
Score = 322 bits (826), Expect = 6e-88, Method: Compositional matrix adjust.
Identities = 140/219 (63%), Positives = 185/219 (84%), Gaps = 0/219 (0%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LWFAILQ+QIETG PYMLYKD+CN KSN ++LGT++ SNLCCE+++YTSP+EVAVCNL
Sbjct 426 QDLWFAILQSQIETGVPYMLYKDSCNAKSNQKNLGTIKCSNLCCEIIEYTSPDEVAVCNL 485
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGV 124
AS++L +FVD EKK F++ KL ++ K++TRNL+ II+RNYYP+ EA+ SN RHRPIG+GV
Sbjct 486 ASIALCKFVDLEKKEFNFKKLYEITKIITRNLDKIIERNYYPVKEAKTSNTRHRPIGIGV 545
Query 125 QGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGV 184
QGLAD F+LLR P++S AA+++NK IFE +Y+AA E S+ELA+IHG YE+Y+GSP S+G+
Sbjct 546 QGLADTFMLLRYPYESDAAKELNKRIFETMYYAALEMSVELASIHGPYESYQGSPASQGI 605
Query 185 FQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
QFDMW K D+ DW L+ K++K+GLRNSLL++PMP
Sbjct 606 LQFDMWNAKVDNKYWDWDELKAKIRKHGLRNSLLLAPMP 644
> tpv:TP03_0528 ribonucleotide-diphosphate reductase large subunit
(EC:1.17.4.1); K10807 ribonucleoside-diphosphate reductase
subunit M1 [EC:1.17.4.1]
Length=898
Score = 318 bits (814), Expect = 1e-86, Method: Compositional matrix adjust.
Identities = 139/219 (63%), Positives = 181/219 (82%), Gaps = 0/219 (0%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LWFAILQ+QIETGTPYMLYKDACN KSN Q+LGT++SSNLCCE+VQ+T+ +EVAVCNL
Sbjct 444 QKLWFAILQSQIETGTPYMLYKDACNAKSNQQNLGTIKSSNLCCEIVQFTNKDEVAVCNL 503
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGV 124
ASV+LP+FV+ + +TFD+ KL ++ +V+T NLN +IDRNYYP+ +AR SN RHRP+G+GV
Sbjct 504 ASVALPKFVNTQTRTFDFKKLYEICRVITYNLNKVIDRNYYPVKQARVSNFRHRPMGVGV 563
Query 125 QGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGV 184
QGLAD F+L+R PF+S AR++NK IFE +Y+A S++LA GTYETYEGSP SKG+
Sbjct 564 QGLADTFMLMRYPFESDEARELNKRIFETMYYACLSESIDLARQLGTYETYEGSPASKGL 623
Query 185 FQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
QFD+WG K D+ L DW L+ ++++GLRNSL ++PMP
Sbjct 624 LQFDLWGAKVDNSLWDWDKLKADLREHGLRNSLFIAPMP 662
> sce:YIL066C RNR3, DIN1, RIR3; One of two large regulatory subunits
of ribonucleotide-diphosphate reductase; the RNR complex
catalyzes rate-limiting step in dNTP synthesis, regulated
by DNA replication and DNA damage checkpoint pathways via
localization of small subunits (EC:1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=869
Score = 311 bits (796), Expect = 2e-84, Method: Compositional matrix adjust.
Identities = 137/225 (60%), Positives = 180/225 (80%), Gaps = 5/225 (2%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW+AILQAQ ETGTP+M+YKDACNRK+N Q+LGT++SSNLCCE+V+Y+SP+E AVC
Sbjct 384 KAQKLWYAILQAQTETGTPFMVYKDACNRKTNQQNLGTIKSSNLCCEIVEYSSPDETAVC 443
Query 63 NLASVSLPRFV----DAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHR 118
NLAS++LP FV D + ++++++L ++ KV+T NLN +IDRNYYP+PEAR SN++HR
Sbjct 444 NLASIALPAFVEVSEDGKTASYNFERLHEIAKVITHNLNRVIDRNYYPVPEARNSNMKHR 503
Query 119 PIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGS 178
PI LGVQGLAD +++LRLPF+S A+ +NK IFE IY A EAS ELA G Y T+EGS
Sbjct 504 PIALGVQGLADTYMMLRLPFESEEAQTLNKQIFETIYHATLEASCELAQKEGKYSTFEGS 563
Query 179 PMSKGVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
P SKG+ QFDMW KP G+ DW+ LR+ + K+GLRNSL ++PMP
Sbjct 564 PASKGILQFDMWNAKP-FGMWDWETLRKDIVKHGLRNSLTMAPMP 607
> mmu:20133 Rrm1, RnrM1; ribonucleotide reductase M1 (EC:1.17.4.1);
K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=792
Score = 308 bits (790), Expect = 7e-84, Method: Compositional matrix adjust.
Identities = 142/221 (64%), Positives = 181/221 (81%), Gaps = 2/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW+AI+++Q ETGTPYMLYKD+CNRKSN Q+LGT++ SNLC E+V+YTS +EVAVC
Sbjct 385 KAQQLWYAIIESQTETGTPYMLYKDSCNRKSNQQNLGTIKCSNLCTEIVEYTSKDEVAVC 444
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS++L +V E T+D++KL +V KV+ RNLN IID NYYPIPEA SN RHRPIG+
Sbjct 445 NLASLALNMYVTPEH-TYDFEKLAEVTKVIVRNLNKIIDINYYPIPEAHLSNKRHRPIGI 503
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
GVQGLADAF+L+R PF+SP A+ +NK IFE IY+ A EAS ELA +G YETYEGSP+SK
Sbjct 504 GVQGLADAFILMRYPFESPEAQLLNKQIFETIYYGALEASCELAKEYGPYETYEGSPVSK 563
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ Q+DMW V P + L DW+ L++K+ KYG+RNSLL++PMP
Sbjct 564 GILQYDMWNVAP-TDLWDWKPLKEKIAKYGIRNSLLIAPMP 603
> cel:T23G5.1 rnr-1; RiboNucleotide Reductase family member (rnr-1);
K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=788
Score = 305 bits (780), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 141/217 (64%), Positives = 175/217 (80%), Gaps = 2/217 (0%)
Query 7 LWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNLAS 66
LW I+ QIETG PY+ YKDA NRKSN Q+LGT++ SNLC E+++Y++P+E+AVCNLAS
Sbjct 395 LWEHIVSNQIETGLPYITYKDAANRKSNQQNLGTIKCSNLCTEIIEYSAPDEIAVCNLAS 454
Query 67 VSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGVQG 126
++L R+V EKK FD+ KL +V KV+TRNLN IID NYYP+ EAR SN+RHRPIGLGVQG
Sbjct 455 IALNRYVTPEKK-FDFVKLAEVTKVITRNLNKIIDVNYYPVEEARNSNMRHRPIGLGVQG 513
Query 127 LADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGVFQ 186
LAD F+L+R PF S ARD+NK IFE IY+AA EAS ELA ++G Y TYEGSP+SKG Q
Sbjct 514 LADCFMLMRYPFTSAEARDLNKRIFETIYYAALEASCELAELNGPYSTYEGSPVSKGQLQ 573
Query 187 FDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
FDMWGV P + CDW LR+K+ K+G+RNSLL++PMP
Sbjct 574 FDMWGVTP-TDQCDWATLRKKIAKHGIRNSLLMAPMP 609
> sce:YER070W RNR1, CRT7, RIR1, SDS12; One of two large regulatory
subunits of ribonucleotide-diphosphate reductase; the RNR
complex catalyzes rate-limiting step in dNTP synthesis, regulated
by DNA replication and DNA damage checkpoint pathways
via localization of small subunits (EC:1.17.4.1); K10807
ribonucleoside-diphosphate reductase subunit M1 [EC:1.17.4.1]
Length=888
Score = 302 bits (774), Expect = 6e-82, Method: Compositional matrix adjust.
Identities = 136/225 (60%), Positives = 179/225 (79%), Gaps = 5/225 (2%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW++IL+AQ ETGTP+++YKDACNRKSN ++LG ++SSNLCCE+V+Y++P+E AVC
Sbjct 384 KAQKLWYSILEAQTETGTPFVVYKDACNRKSNQKNLGVIKSSNLCCEIVEYSAPDETAVC 443
Query 63 NLASVSLPRFV----DAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHR 118
NLASV+LP F+ D + T+++ KL ++ KV+TRNLN +IDRNYYP+ EAR+SN+RHR
Sbjct 444 NLASVALPAFIETSEDGKTSTYNFKKLHEIAKVVTRNLNRVIDRNYYPVEEARKSNMRHR 503
Query 119 PIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGS 178
PI LGVQGLAD F+LLRLPFDS AR +N IFE IY A+ EAS ELA G YET++GS
Sbjct 504 PIALGVQGLADTFMLLRLPFDSEEARLLNIQIFETIYHASMEASCELAQKDGPYETFQGS 563
Query 179 PMSKGVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
P S+G+ QFDMW KP G+ DW LR+ + K+G+RNSL ++PMP
Sbjct 564 PASQGILQFDMWDQKP-YGMWDWDTLRKDIMKHGVRNSLTMAPMP 607
> hsa:6240 RRM1, R1, RIR1, RR1; ribonucleotide reductase M1 (EC:1.17.4.1);
K10807 ribonucleoside-diphosphate reductase subunit
M1 [EC:1.17.4.1]
Length=792
Score = 301 bits (771), Expect = 1e-81, Method: Compositional matrix adjust.
Identities = 139/221 (62%), Positives = 179/221 (80%), Gaps = 2/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW+AI+++Q ETGTPYMLYKD+CNRKSN Q+LGT++ SNLC E+V+YTS +EVAVC
Sbjct 385 KAQQLWYAIIESQTETGTPYMLYKDSCNRKSNQQNLGTIKCSNLCTEIVEYTSKDEVAVC 444
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS++L +V +E T+D+ KL +V KV+ RNLN IID NYYP+PEA SN RHRPIG+
Sbjct 445 NLASLALNMYVTSEH-TYDFKKLAEVTKVVVRNLNKIIDINYYPVPEACLSNKRHRPIGI 503
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
GVQGLADAF+L+R PF+S A+ +NK IFE IY+ A EAS +LA G YETYEGSP+SK
Sbjct 504 GVQGLADAFILMRYPFESAEAQLLNKQIFETIYYGALEASCDLAKEQGPYETYEGSPVSK 563
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ Q+DMW V P + L DW+ L++K+ KYG+RNSLL++PMP
Sbjct 564 GILQYDMWNVTP-TDLWDWKVLKEKIAKYGIRNSLLIAPMP 603
> xla:399431 rrm1; ribonucleotide reductase M1 (EC:1.17.4.1);
K10807 ribonucleoside-diphosphate reductase subunit M1 [EC:1.17.4.1]
Length=797
Score = 296 bits (759), Expect = 3e-80, Method: Compositional matrix adjust.
Identities = 137/221 (61%), Positives = 177/221 (80%), Gaps = 2/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q +W AI+++Q ETGTPYMLYKDACNRKSN Q+LGT++ SNLC E+V+YTS +EVAVC
Sbjct 385 KAQQVWHAIIESQTETGTPYMLYKDACNRKSNQQNLGTIKCSNLCTEIVEYTSDKEVAVC 444
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS++L +V E+ +FD+ KL DV KV+ RNLN II+ N+YP+PEA SN +HRPIG+
Sbjct 445 NLASLALNMYVTPER-SFDFKKLADVTKVIVRNLNKIIEINFYPVPEAEYSNKKHRPIGI 503
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
GVQGLADAF+L+R PF+S A+ +NK IFE IY+AA E+S ELA G YETYEG P+SK
Sbjct 504 GVQGLADAFILMRYPFESEKAQMLNKQIFETIYYAALESSCELAKELGPYETYEGCPVSK 563
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ Q+DMW V P + L DW L++K+ KYG+RNSLL++PMP
Sbjct 564 GILQYDMWNVTP-TDLWDWTALKEKIAKYGIRNSLLLAPMP 603
> ath:AT2G21790 RNR1; RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ATP binding
/ protein binding / ribonucleoside-diphosphate reductase
(EC:1.17.4.1); K10807 ribonucleoside-diphosphate reductase
subunit M1 [EC:1.17.4.1]
Length=816
Score = 292 bits (747), Expect = 7e-79, Method: Compositional matrix adjust.
Identities = 139/237 (58%), Positives = 175/237 (73%), Gaps = 19/237 (8%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LW+ IL +Q+ETGTPYML+KD+CNRKSN Q+LGT++SSNLC E+++YTSP E AVCNL
Sbjct 387 QQLWYEILTSQVETGTPYMLFKDSCNRKSNQQNLGTIKSSNLCTEIIEYTSPTETAVCNL 446
Query 65 ASVSLPRFV------------------DAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYP 106
AS++LPRFV D++ + FD++KL +V +T NLN IID NYYP
Sbjct 447 ASIALPRFVREKGVPLDSHPPKLAGSLDSKNRYFDFEKLAEVTATVTVNLNKIIDVNYYP 506
Query 107 IPEARRSNLRHRPIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELA 166
+ A+ SN+RHRPIG+GVQGLADAF+LL +PFDSP A+ +NK+IFE IY+ A +AS ELA
Sbjct 507 VETAKTSNMRHRPIGIGVQGLADAFILLGMPFDSPEAQQLNKDIFETIYYHALKASTELA 566
Query 167 AIHGTYETYEGSPMSKGVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
A G YETY GSP+SKG+ Q DMW V P S DW LR + K G+RNSLLV+PMP
Sbjct 567 ARLGPYETYAGSPVSKGILQPDMWNVIP-SDRWDWAVLRDMISKNGVRNSLLVAPMP 622
> dre:30740 rrm1, CHUNP6866, cb396, cb838, r1, sb:cb548, wu:fb39b07,
wu:fi14b02, wu:fk95f07; ribonucleotide reductase M1 polypeptide
(EC:1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=794
Score = 291 bits (744), Expect = 2e-78, Method: Compositional matrix adjust.
Identities = 134/221 (60%), Positives = 174/221 (78%), Gaps = 2/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW+AI+++Q ETGTPYMLYKDACNRKSN Q+LGT++ SNLC E+V+YTS +EVAVC
Sbjct 385 KAQQLWYAIIESQTETGTPYMLYKDACNRKSNRQNLGTIKCSNLCTEIVEYTSADEVAVC 444
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS++L +V +E+ TFD+ KL V KV+ +NLN IID NYYP+ EA SN RHRPIG+
Sbjct 445 NLASIALNMYVTSER-TFDFQKLASVTKVIVKNLNKIIDINYYPVKEAENSNKRHRPIGI 503
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
GVQGLADAF+L+R PF+S A+ +N IFE IY+AA E+S ELAA +G Y+TY G P+SK
Sbjct 504 GVQGLADAFILMRFPFESTEAQLLNTQIFETIYYAALESSCELAAEYGPYQTYAGCPVSK 563
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ Q+DMW P + L DW L++K+ G+RNSLL++PMP
Sbjct 564 GILQYDMWEKTP-TDLWDWAALKEKIANDGVRNSLLLAPMP 603
> eco:b2234 nrdA, dnaF, ECK2226, JW2228; ribonucleoside-diphosphate
reductase 1, alpha subunit (EC:1.17.4.1); K00525 ribonucleoside-diphosphate
reductase alpha chain [EC:1.17.4.1]
Length=761
Score = 93.6 bits (231), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 65/231 (28%), Positives = 108/231 (46%), Gaps = 21/231 (9%)
Query 7 LWFAILQAQIETGTPYMLYKDACNRKSNHQ-HLGTLRSSNLCCEVVQYTSP--------E 57
L+ ++Q + TG Y+ D CN S + +R SNLC E+ T P
Sbjct 398 LFSLMMQERASTGRIYIQNVDHCNTHSPFDPAIAPVRQSNLCLEIALPTKPLNDVNDENG 457
Query 58 EVAVCNLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRH 117
E+A+C L++ +L + D+L+++ + R L+ ++D YPIP A+R +
Sbjct 458 EIALCTLSAFNL-------GAINNLDELEELAILAVRALDALLDYQDYPIPAAKRGAMGR 510
Query 118 RPIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEG 177
R +G+GV A + +A ++ FE I + +AS ELA G +
Sbjct 511 RTLGIGVINFAYYLAKHGKRYSDGSANNLTHKTFEAIQYYLLKASNELAKEQGACPWFNE 570
Query 178 SPMSKGVFQFDMWGVKPDSGL-----CDWQGLRQKVKKYGLRNSLLVSPMP 223
+ +KG+ D + D+ DW+ LR+ +K +GLRNS L + MP
Sbjct 571 TTYAKGILPIDTYKKDLDTIANEPLHYDWEALRESIKTHGLRNSTLSALMP 621
> eco:b2675 nrdE, ECK2669, JW2650; ribonucleoside-diphosphate
reductase 2, alpha subunit (EC:1.17.4.1); K00525 ribonucleoside-diphosphate
reductase alpha chain [EC:1.17.4.1]
Length=714
Score = 63.5 bits (153), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 60/244 (24%), Positives = 93/244 (38%), Gaps = 40/244 (16%)
Query 11 ILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEV----------- 59
+ + Q E+G PY++Y+D NR + G + SNLC E++Q S E
Sbjct 354 LAEIQFESGYPYIMYEDTVNRA--NPIAGRINMSNLCSEILQVNSASEYDENLDYTRTGH 411
Query 60 -AVCNLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYY-PIPEARRSNLRH 117
CNL S+++ T D ++ R L + D ++ +P N
Sbjct 412 DISCNLGSLNIAH-------TMDSPDFARTVETAVRGLTAVSDMSHIRSVPSIEAGNAAS 464
Query 118 RPIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHG-TYETYE 176
IGLG L + + SP A D F I + A SM LA G T+ ++
Sbjct 465 HAIGLGQMNLHGYLAREGIAYGSPEALDFTNLYFYAITWHALRTSMLLARERGETFAGFK 524
Query 177 GSPMSKGVF--QFDMWGVKPDSGLCD---------------WQGLRQKVKKYGLRNSLLV 219
S + G + Q+ +P + W LR V +YG+ N L
Sbjct 525 QSRYASGEYFSQYLQGNWQPKTAKVGELFTRSGITLPTREMWAQLRDDVMRYGIYNQNLQ 584
Query 220 SPMP 223
+ P
Sbjct 585 AVPP 588
> ath:AT1G53020 UBC26; UBC26 (UBIQUITIN-CONJUGATING ENZYME 26);
ubiquitin-protein ligase
Length=1163
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 28/46 (60%), Gaps = 6/46 (13%)
Query 146 INKNIFECIYFAACEASMEL--AAIHGTYETYEGSPMSKGVFQFDM 189
+N+N+ E I+ ACE+ M+L A I G EG+P G+F FD+
Sbjct 929 LNQNLPETIFVRACESRMDLLRAVIIGA----EGTPYHDGLFFFDI 970
Score = 32.7 bits (73), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 6/46 (13%)
Query 146 INKNIFECIYFAACEASMEL--AAIHGTYETYEGSPMSKGVFQFDM 189
++KN+ E I+ ACE+ ++L A I G EG+P G+F FD+
Sbjct 617 LDKNLPETIFVRACESRIDLLRAVIIGA----EGTPYHDGLFFFDI 658
Lambda K H
0.322 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7395169300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40