bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1112_orf1
Length=320
Score E
Sequences producing significant alignments: (Bits) Value
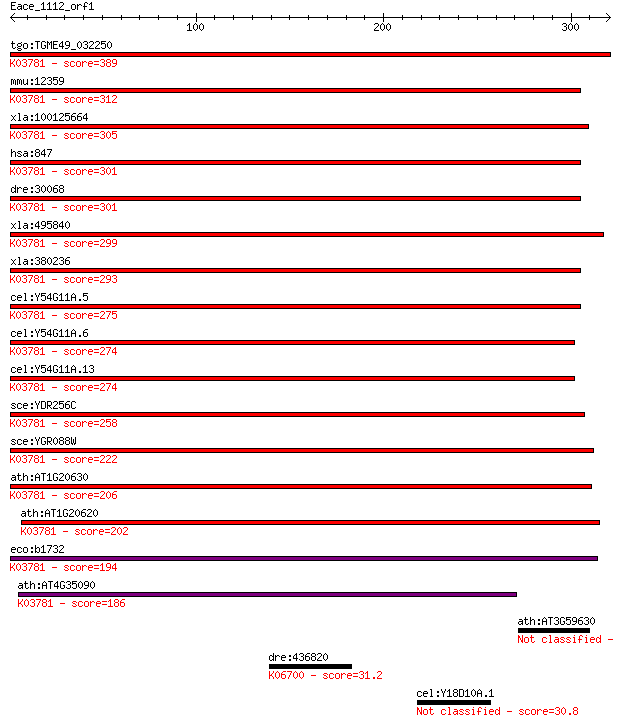
tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781 c... 389 9e-108
mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.1... 312 1e-84
xla:100125664 hypothetical protein LOC100125664; K03781 catala... 305 1e-82
hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03... 301 2e-81
dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6); K0... 301 3e-81
xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 cata... 299 1e-80
xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catal... 293 6e-79
cel:Y54G11A.5 ctl-2; CaTaLase family member (ctl-2); K03781 ca... 275 2e-73
cel:Y54G11A.6 ctl-1; CaTaLase family member (ctl-1); K03781 ca... 274 4e-73
cel:Y54G11A.13 ctl-3; CaTaLase family member (ctl-3); K03781 c... 274 4e-73
sce:YDR256C CTA1; Catalase A, breaks down hydrogen peroxide in... 258 1e-68
sce:YGR088W CTT1, SPS101; Ctt1p (EC:1.11.1.6); K03781 catalase... 222 1e-57
ath:AT1G20630 CAT1; CAT1 (CATALASE 1); catalase (EC:1.11.1.6);... 206 9e-53
ath:AT1G20620 CAT3; CAT3 (CATALASE 3); catalase; K03781 catala... 202 1e-51
eco:b1732 katE, ECK1730, JW1721; hydroperoxidase HPII(III) (ca... 194 4e-49
ath:AT4G35090 CAT2; CAT2 (CATALASE 2); catalase (EC:1.11.1.6);... 186 8e-47
ath:AT3G59630 diphthamide synthesis DPH2 family protein 32.7 2.1
dre:436820 psmf1, zgc:92785; proteasome (prosome, macropain) i... 31.2 5.1
cel:Y18D10A.1 hypothetical protein 30.8 6.2
> tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=502
Score = 389 bits (999), Expect = 9e-108, Method: Compositional matrix adjust.
Identities = 177/321 (55%), Positives = 235/321 (73%), Gaps = 1/321 (0%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
VHQ T L++DRGTPDG+RHMNGY SH FK +N +NE FY K+H+KTNQGIKNL+ + A+
Sbjct 182 VHQVTFLYTDRGTPDGFRHMNGYGSHTFKFINKDNEAFYVKWHFKTNQGIKNLNRQRAKE 241
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
L DPDYA RDLFNAI + +FPSWT IQVMP+++A Y++N+ DVTKV PH DYPL
Sbjct 242 LESEDPDYAVRDLFNAIAKREFPSWTFCIQVMPLKDAETYKWNVFDVTKVWPHGDYPLIP 301
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VGRL+L+ NP+NYFQDVEQ+AF+P+H++PG+EPSED+MLQGR+F+Y D HRHRLG NY Q
Sbjct 302 VGRLVLDRNPENYFQDVEQAAFAPAHMVPGIEPSEDRMLQGRMFSYIDTHRHRLGANYHQ 361
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKE-GLHSSTPLHGST 239
IP+N P + +RDG MCV+GN NY P+ +E + + +T + G+
Sbjct 362 IPVNRPWNARGGDYSVRDGPMCVDGNKGSQLNYEPNSVDGFPKEDRNAAVSGTTTVSGTV 421
Query 240 GHHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFARADED 299
H Q+HPNSDFEQ G YR V+++ +R+ LI NI L+ A R++QERQVK+F + D +
Sbjct 422 ACHPQEHPNSDFEQPGNFYRTVLSEPEREALIGNIAEHLRQARRDIQERQVKIFYKCDPE 481
Query 300 YGRRVAKGLGLPLSVCSSSRM 320
YG RVA+ +GLP + C ++M
Sbjct 482 YGERVARAIGLPTAACYPAKM 502
> mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=527
Score = 312 bits (799), Expect = 1e-84, Method: Compositional matrix adjust.
Identities = 161/305 (52%), Positives = 198/305 (64%), Gaps = 5/305 (1%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ + LFSDRG PDG+RHMNGY SH FK VN++ E Y KFH+KT+QGIKNL EA R
Sbjct 193 LHQVSFLFSDRGIPDGHRHMNGYGSHTFKLVNADGEAVYCKFHYKTDQGIKNLPVGEAGR 252
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA DPDY RDLFNAI G++PSWT YIQVM +EA + +N D+TKV PHKDYPL
Sbjct 253 LAQEDPDYGLRDLFNAIANGNYPSWTFYIQVMTFKEAETFPFNPFDLTKVWPHKDYPLIP 312
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+L+LN NP NYF +VEQ AF PS++ PG+EPS DKMLQGRLFAYPD HRHRLGPNY Q
Sbjct 313 VGKLVLNKNPVNYFAEVEQMAFDPSNMPPGIEPSPDKMLQGRLFAYPDTHRHRLGPNYLQ 372
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKEGLHSSTPLHGSTG 240
IP+N P + A Y RDG MC++ N NY P+ + ++ HS + +
Sbjct 373 IPVNCPYRARV-ANYQRDGPMCMHDNQGGAPNYYPNSFSAPEQQRSALEHS---VQCAVD 428
Query 241 HHRQQHPNSD-FEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFARADED 299
R N D QV Y +V+N+ +R L NI LK A +Q++ VK F D
Sbjct 429 VKRFNSANEDNVTQVRTFYTKVLNEEERKRLCENIAGHLKDAQLFIQKKAVKNFTDVHPD 488
Query 300 YGRRV 304
YG R+
Sbjct 489 YGARI 493
> xla:100125664 hypothetical protein LOC100125664; K03781 catalase
[EC:1.11.1.6]
Length=503
Score = 305 bits (781), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 156/308 (50%), Positives = 198/308 (64%), Gaps = 3/308 (0%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ T LFSDRG PDG+RHMNGY SH FK VN+E Y KFH+KT+QGIKNLS EEA+R
Sbjct 175 LHQVTFLFSDRGIPDGHRHMNGYGSHTFKLVNAEGNAVYCKFHYKTDQGIKNLSVEEADR 234
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
L +DPDY RDLF AI + +FPSWT+YIQVM EA +N D+TKV PHKDYPL
Sbjct 235 LVVSDPDYGIRDLFQAIAKKNFPSWTMYIQVMTFEEAEKCPFNPFDLTKVWPHKDYPLIP 294
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
+G+L+LN NP+NYF +VEQ AF PSH+ PG+E S DKMLQGRLF+YPD HR+RLGPNY
Sbjct 295 IGKLVLNRNPENYFAEVEQIAFDPSHMPPGIEASPDKMLQGRLFSYPDTHRYRLGPNYLH 354
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKEGLHSSTPLHGSTG 240
+P+N PRG + A Y RDG MC+ + + NY P+ +S R+ + S+ G
Sbjct 355 LPVNCPRGVKV-AHYQRDGPMCMFNTPSHMPNYYPN-SFSSPRDDPKCKDSTFVASGDVD 412
Query 241 HHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFARADEDY 300
H + QV Y + + + +R L N+ +L A +QER VK F+ DY
Sbjct 413 RHDCSD-EDNVSQVRAFYTQTLTEEERQRLCENLARNLSEAQIFIQERAVKNFSNIHPDY 471
Query 301 GRRVAKGL 308
G + L
Sbjct 472 GADIKALL 479
> hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=527
Score = 301 bits (771), Expect = 2e-81, Method: Compositional matrix adjust.
Identities = 159/305 (52%), Positives = 190/305 (62%), Gaps = 5/305 (1%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ + LFSDRG PDG+RHMNGY SH FK VN+ E Y KFH+KT+QGIKNLS E+A R
Sbjct 193 LHQVSFLFSDRGIPDGHRHMNGYGSHTFKLVNANGEAVYCKFHYKTDQGIKNLSVEDAAR 252
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
L+ DPDY RDLFNAI G +PSWT YIQVM +A + +N D+TKV PHKDYPL
Sbjct 253 LSQEDPDYGIRDLFNAIATGKYPSWTFYIQVMTFNQAETFPFNPFDLTKVWPHKDYPLIP 312
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+L+LN NP NYF +VEQ AF PS++ PG+E S DKMLQGRLFAYPD HRHRLGPNY
Sbjct 313 VGKLVLNRNPVNYFAEVEQIAFDPSNMPPGIEASPDKMLQGRLFAYPDTHRHRLGPNYLH 372
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKEGLHSSTPLHGSTG 240
IP+N P + A Y RDG MC+ N NY P+ + + L S G
Sbjct 373 IPVNCPYRARV-ANYQRDGPMCMQDNQGGAPNYYPN-SFGAPEQQPSALEHSIQYSGEV- 429
Query 241 HHRQQHPNSD-FEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFARADED 299
R N D QV Y V+N+ QR L NI LK A +Q++ VK F D
Sbjct 430 -RRFNTANDDNVTQVRAFYVNVLNEEQRKRLCENIAGHLKDAQIFIQKKAVKNFTEVHPD 488
Query 300 YGRRV 304
YG +
Sbjct 489 YGSHI 493
> dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=526
Score = 301 bits (770), Expect = 3e-81, Method: Compositional matrix adjust.
Identities = 152/304 (50%), Positives = 198/304 (65%), Gaps = 3/304 (0%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ + LFSDRG PDGYRHMNGY SH FK VN++ + Y KFH+KTNQGIKN+ EEA+R
Sbjct 193 LHQVSFLFSDRGIPDGYRHMNGYGSHTFKLVNAQGQPVYCKFHYKTNQGIKNIPVEEADR 252
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA TDPDY+ RDL+NAI G+FPSWT YIQVM +A ++++N D+TKV HK++PL
Sbjct 253 LAATDPDYSIRDLYNAIANGNFPSWTFYIQVMTFEQAENWKWNPFDLTKVWSHKEFPLIP 312
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VGR +LN NP NYF +VEQ AF PS++ PG+EPS DKMLQGRLF+YPD HRHRLG NY Q
Sbjct 313 VGRFVLNRNPVNYFAEVEQLAFDPSNMPPGIEPSPDKMLQGRLFSYPDTHRHRLGANYLQ 372
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKEGLHSSTPLHGSTG 240
+P+N P + A Y RDG MC++ N NY P+ + S +V+ S
Sbjct 373 LPVNCPYRTRV-ANYQRDGPMCMHDNQGGAPNYYPN--SFSAPDVQPRFLESKCKVSPDV 429
Query 241 HHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFARADEDY 300
+ + QV + +V+N+ +R+ L N+ LK A +Q+R V+ DY
Sbjct 430 ARYNSADDDNVTQVRTFFTQVLNEAERERLCQNMAGHLKGAQLFIQKRMVQNLMAVHSDY 489
Query 301 GRRV 304
G RV
Sbjct 490 GNRV 493
> xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 299 bits (765), Expect = 1e-80, Method: Compositional matrix adjust.
Identities = 153/321 (47%), Positives = 202/321 (62%), Gaps = 13/321 (4%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ + LFSDRG PDG+RHMNGY SH FK V+ ++E Y KFH+KT+QGI+NL+ E AE+
Sbjct 193 LHQVSFLFSDRGIPDGHRHMNGYGSHTFKLVSCKDEAVYCKFHFKTDQGIRNLTVERAEQ 252
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA +DPDY RDL+ AI G++PSWTLYIQ M ++A + +N DVTKV PH DYPL
Sbjct 253 LAASDPDYGIRDLYEAIAAGNYPSWTLYIQTMTFQQAEKFPFNPFDVTKVWPHGDYPLIP 312
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+++L+ NPKNYF +VEQ AF PS++ PG+EPS DKMLQGRLF+YPD HRHRLG NY Q
Sbjct 313 VGKMVLSQNPKNYFAEVEQLAFDPSNMPPGIEPSPDKMLQGRLFSYPDTHRHRLGTNYLQ 372
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPS-----EETNSLREVKEGLHSSTPL 235
+PIN P + A Y RDG MC N NY P+ E+ +E K + +
Sbjct 373 LPINCPYKVRV-ANYQRDGPMCFTDNQGGAPNYYPNSFSAPEQQPQFKEHKFRVSADVDR 431
Query 236 HGSTGHHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFAR 295
+ S + + QV + Y V+N+ +R L N+ LK +Q+R VK F+
Sbjct 432 YNSAV-------DDNVTQVRDFYLNVLNEEERQRLCENLAGHLKECQLFIQKRAVKNFSD 484
Query 296 ADEDYGRRVAKGLGLPLSVCS 316
DYG R+ L + C+
Sbjct 485 VHLDYGSRIQALLDKHNAKCT 505
> xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 293 bits (750), Expect = 6e-79, Method: Compositional matrix adjust.
Identities = 148/309 (47%), Positives = 197/309 (63%), Gaps = 13/309 (4%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ + LFSDRG PDG+RHMNGY SH FK VN+++E Y KFH+KT+Q I+NL+ +EA R
Sbjct 193 LHQVSFLFSDRGIPDGHRHMNGYGSHTFKLVNAKDEAVYCKFHYKTDQCIQNLTVDEANR 252
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA +DPDY DL+ AI G++PSW+ YIQVM +A +++N D+TK+ PH DYPL
Sbjct 253 LAASDPDYGIHDLYEAITTGNYPSWSFYIQVMTFEQAERFKFNPFDLTKIWPHGDYPLIP 312
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+L+LN NP NYF +VEQ AF PS++ PG+EPS DKMLQGRLF+YPD HRHRLGPNY Q
Sbjct 313 VGKLVLNRNPTNYFAEVEQLAFDPSNMPPGIEPSPDKMLQGRLFSYPDTHRHRLGPNYLQ 372
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPS-----EETNSLREVKEGLHSSTPL 235
+P+N P + A Y RD MC N NY P+ E +RE + + +
Sbjct 373 LPVNCPYRTRV-ANYQRDRPMCFTDNQGGAPNYYPNSFCAPENQPQVREHRFQVSADVAR 431
Query 236 HGSTGHHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFAR 295
+ S+ + QV + Y +V+++ QR L NI LK A +Q+R VK F
Sbjct 432 YNSSDE-------DNVSQVRDFYVKVLSEEQRLRLCENIAGHLKDAQLFIQKRAVKNFTD 484
Query 296 ADEDYGRRV 304
+YG R+
Sbjct 485 VHPEYGARI 493
> cel:Y54G11A.5 ctl-2; CaTaLase family member (ctl-2); K03781
catalase [EC:1.11.1.6]
Length=500
Score = 275 bits (702), Expect = 2e-73, Method: Compositional matrix adjust.
Identities = 137/304 (45%), Positives = 190/304 (62%), Gaps = 4/304 (1%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ LFSDRG PDGYRHMNGY SH FK VN + + Y KFH+K QG+KNL+ E+A +
Sbjct 189 LHQVMFLFSDRGLPDGYRHMNGYGSHTFKMVNKDGKAIYVKFHFKPTQGVKNLTVEKAGQ 248
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA +DPDY+ RDLFNAIE+GDFP W ++IQVM +A + +N DVTKV PH DYPL
Sbjct 249 LASSDPDYSIRDLFNAIEKGDFPVWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIE 308
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+++LN NP+NYF +VEQSAF P+H++PG+E S DKMLQGR+F+Y D H HRLGPNY Q
Sbjct 309 VGKMVLNRNPRNYFAEVEQSAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHFHRLGPNYIQ 368
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKEGLHSSTPLHGSTG 240
+P+N P H RDG M + N N+ P+ N + + ++ P G
Sbjct 369 LPVNCPYRSRAHN-TQRDGAMAYD-NQQHAPNFFPN-SFNYGKTRPDVKDTTFPATGDVD 425
Query 241 HHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFARADEDY 300
+ N +++Q + + +V++ R+ + N L + + + F++ D+
Sbjct 426 RYESGDDN-NYDQPRQFWEKVLDTGARERMCQNFAGPLGECHDFIIKGMIDHFSKVHPDF 484
Query 301 GRRV 304
G RV
Sbjct 485 GARV 488
> cel:Y54G11A.6 ctl-1; CaTaLase family member (ctl-1); K03781
catalase [EC:1.11.1.6]
Length=497
Score = 274 bits (700), Expect = 4e-73, Method: Compositional matrix adjust.
Identities = 142/307 (46%), Positives = 187/307 (60%), Gaps = 16/307 (5%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ L+SDRG PDG+R MNGY +H FK VN E Y KFH+K QG KNL +A +
Sbjct 189 IHQVMFLYSDRGIPDGFRFMNGYGAHTFKMVNKEGNPIYCKFHFKPAQGSKNLDPTDAGK 248
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA +DPDYA RDLFNAIE +FP W ++IQVM +A + +N DVTKV PH DYPL
Sbjct 249 LASSDPDYAIRDLFNAIESRNFPEWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIE 308
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+++LN N KNYF +VEQ+AF P+H++PG+E S DKMLQGR+F+Y D H HRLGPNY Q
Sbjct 309 VGKMVLNRNVKNYFAEVEQAAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHYHRLGPNYIQ 368
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLR------EVKEGLHSSTP 234
+P+N P H RDG M D NY P NS R +VKE +T
Sbjct 369 LPVNCPYRSRAHT-TQRDGAMAYESQ-GDAPNYFP----NSFRGYRTRDDVKESTFQTT- 421
Query 235 LHGSTGHHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFA 294
G + + + ++EQ + + +V+ + +RD L+ N+ S L E+Q VK F
Sbjct 422 --GDVDRY-ETGDDHNYEQPRQFWEKVLKEEERDRLVGNLASDLGGCLEEIQNGMVKEFT 478
Query 295 RADEDYG 301
+ D+G
Sbjct 479 KVHPDFG 485
> cel:Y54G11A.13 ctl-3; CaTaLase family member (ctl-3); K03781
catalase [EC:1.11.1.6]
Length=512
Score = 274 bits (700), Expect = 4e-73, Method: Compositional matrix adjust.
Identities = 142/307 (46%), Positives = 187/307 (60%), Gaps = 16/307 (5%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ L+SDRG PDG+R MNGY +H FK VN E Y KFH+K QG KNL +A +
Sbjct 204 IHQVMFLYSDRGIPDGFRFMNGYGAHTFKMVNKEGNPIYCKFHFKPAQGSKNLDPTDAGK 263
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
LA +DPDYA RDLFNAIE +FP W ++IQVM +A + +N DVTKV PH DYPL
Sbjct 264 LASSDPDYAIRDLFNAIESRNFPEWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIE 323
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+++LN N KNYF +VEQ+AF P+H++PG+E S DKMLQGR+F+Y D H HRLGPNY Q
Sbjct 324 VGKMVLNRNVKNYFAEVEQAAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHYHRLGPNYIQ 383
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLR------EVKEGLHSSTP 234
+P+N P H RDG M D NY P NS R +VKE +T
Sbjct 384 LPVNCPYRSRAHTTQ-RDGAMAYESQ-GDAPNYFP----NSFRGYRTRDDVKESTFQTT- 436
Query 235 LHGSTGHHRQQHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKHASREVQERQVKLFA 294
G + + + ++EQ + + +V+ + +RD L+ N+ S L E+Q VK F
Sbjct 437 --GDVDRY-ETGDDHNYEQPRQFWEKVLKEEERDRLVGNLASDLGGCLEEIQNGMVKEFT 493
Query 295 RADEDYG 301
+ D+G
Sbjct 494 KVHPDFG 500
> sce:YDR256C CTA1; Catalase A, breaks down hydrogen peroxide
in the peroxisomal matrix formed by acyl-CoA oxidase (Pox1p)
during fatty acid beta-oxidation (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=515
Score = 258 bits (660), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 137/315 (43%), Positives = 194/315 (61%), Gaps = 14/315 (4%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ ILFSDRGTP YR M+GYS H +K N + Y + H KT+QGIKNL+ EEA +
Sbjct 190 IHQVMILFSDRGTPANYRSMHGYSGHTYKWSNKNGDWHYVQVHIKTDQGIKNLTIEEATK 249
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
+AG++PDY +DLF AI+ G++PSWT+YIQ M R+A +++ D+TKV P +PLRR
Sbjct 250 IAGSNPDYCQQDLFEAIQNGNYPSWTVYIQTMTERDAKKLPFSVFDLTKVWPQGQFPLRR 309
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VG+++LN NP N+F VEQ+AF+PS +P E S D +LQ RLF+Y DAHR+RLGPN+ Q
Sbjct 310 VGKIVLNENPLNFFAQVEQAAFAPSTTVPYQEASADPVLQARLFSYADAHRYRLGPNFHQ 369
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNY-------ADLKNYTPSEETNSLREVKEGLHS-S 232
IP+N P IRDG M VNGN+ A+ K+YT ++ +++ +E + +
Sbjct 370 IPVNCPYASKFFNPAIRDGPMNVNGNFGSEPTYLANDKSYTYIQQDRPIQQHQEVWNGPA 429
Query 233 TPLHGSTGHHRQQHPNSDFEQVGELYRRVMND-RQRDILITNIVSSLKHASREVQERQVK 291
P H +T + DF Q LYR + Q+ L NI ++ A ++Q+R
Sbjct 430 IPYHWATSPG-----DVDFVQARNLYRVLGKQPGQQKNLAYNIGIHVEGACPQIQQRVYD 484
Query 292 LFARADEDYGRRVAK 306
+FAR D+ + K
Sbjct 485 MFARVDKGLSEAIKK 499
> sce:YGR088W CTT1, SPS101; Ctt1p (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=562
Score = 222 bits (566), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 126/360 (35%), Positives = 189/360 (52%), Gaps = 50/360 (13%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+HQ T +F DRGTP + MN YS H+F VN E + Y +FH ++ G + L+ ++A
Sbjct 185 IHQITYMFGDRGTPASWASMNAYSGHSFIMVNKEGKDTYVQFHVLSDTGFETLTGDKAAE 244
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
L+G+ PDY LF ++ G+ P + Y+Q M +A+ +RY++ D+TK+ PHK++PLR+
Sbjct 245 LSGSHPDYNQAKLFTQLQNGEKPKFNCYVQTMTPEQATKFRYSVNDLTKIWPHKEFPLRK 304
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSH-LIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYD 179
G + L N NYFQ++EQ AFSP++ IPG++PS D +LQ RLF+YPD RHRLG NY
Sbjct 305 FGTITLTENVDNYFQEIEQVAFSPTNTCIPGIKPSNDSVLQARLFSYPDTQRHRLGANYQ 364
Query 180 QIPINVPRG----------------CPLHAF-YIRDGFMCVNGNYADLKNYTPSEETNSL 222
Q+P+N PR CP A + RDG M N+ NY S +L
Sbjct 365 QLPVNRPRNLGCPYSKGDSQYTAEQCPFKAVNFQRDGPMSYY-NFGPEPNYISSLPNQTL 423
Query 223 R-------EVKEGLHSSTPLHGSTGHHRQQHPNS-----------------------DFE 252
+ EV + + R+Q + DFE
Sbjct 424 KFKNEDNDEVSDKFKGIVLDEVTEVSVRKQEQDQIRNEHIVDAKINQYYYVYGISPLDFE 483
Query 253 QVGELYRRVMNDRQRDILITNIVS-SLKHASREVQERQVKLFARADEDYGRRVAKGLGLP 311
Q LY +V ND Q+ + + N+V + K +V++R + F +ED G+ +A+ LG+P
Sbjct 484 QPRALYEKVYNDEQKKLFVHNVVCHACKIKDPKVKKRVTQYFGLLNEDLGKVIAECLGVP 543
> ath:AT1G20630 CAT1; CAT1 (CATALASE 1); catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=492
Score = 206 bits (524), Expect = 9e-53, Method: Compositional matrix adjust.
Identities = 118/314 (37%), Positives = 185/314 (58%), Gaps = 12/314 (3%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+H + LF D G P YRHM G + + +N + Y KFHWK GIK LS EEA R
Sbjct 183 LHMFSFLFDDLGIPQDYRHMEGAGVNTYMLINKAGKAHYVKFHWKPTCGIKCLSDEEAIR 242
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
+ G + +AT+DL+++I G++P W L++QVM + ++ LDVTK+ P PL+
Sbjct 243 VGGANHSHATKDLYDSIAAGNYPQWNLFVQVMDPAHEDKFDFDPLDVTKIWPEDILPLQP 302
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQ 180
VGRL+LN N N+F + EQ AF P+ ++PG+ S+DK+LQ R+F+Y D+ RHRLGPNY Q
Sbjct 303 VGRLVLNKNIDNFFNENEQIAFCPALVVPGIHYSDDKLLQTRIFSYADSQRHRLGPNYLQ 362
Query 181 IPINVPRGCPLHAFYIRDGFMCVNGNYADLK-NYTPSEETNSLREVKEGLHSSTPLHGST 239
+P+N P+ C H + DGFM N + D + NY PS + +R ++ + +TP+ S
Sbjct 363 LPVNAPK-CAHHNNH-HDGFM--NFMHRDEEVNYFPS-RLDPVRHAEK--YPTTPIVCSG 415
Query 240 GHHRQ-QHPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKH--ASREVQERQVKLFARA 296
+ ++F+Q GE YR +DRQ + + V +L + E++ + +++A
Sbjct 416 NREKCFIGKENNFKQPGERYRSWDSDRQ-ERFVKRFVEALSEPRVTHEIRSIWISYWSQA 474
Query 297 DEDYGRRVAKGLGL 310
D+ G+++A L +
Sbjct 475 DKSLGQKLATRLNV 488
> ath:AT1G20620 CAT3; CAT3 (CATALASE 3); catalase; K03781 catalase
[EC:1.11.1.6]
Length=377
Score = 202 bits (515), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 115/312 (36%), Positives = 175/312 (56%), Gaps = 12/312 (3%)
Query 7 LFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAERLAGTDP 66
+F D G P YRHM G+ H + + +V + KFHWK GIKNL+ EEA+ + G +
Sbjct 74 MFDDVGIPQDYRHMEGFGVHTYTLIAKSGKVLFVKFHWKPTCGIKNLTDEEAKVVGGANH 133
Query 67 DYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRRVGRLIL 126
+AT+DL +AI G++P W L+IQ M + + ++ LDVTK+ P PL+ VGRL+L
Sbjct 134 SHATKDLHDAIASGNYPEWKLFIQTMDPADEDKFDFDPLDVTKIWPEDILPLQPVGRLVL 193
Query 127 NVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQIPINVP 186
N N+F + EQ AF+P ++PG+ S+DK+LQ R+FAY D RHRLGPNY Q+P+N P
Sbjct 194 NRTIDNFFNETEQLAFNPGLVVPGIYYSDDKLLQCRIFAYGDTQRHRLGPNYLQLPVNAP 253
Query 187 RGCPLHAFYIRDGFMCVNGNYADLKNYTPSEETNSLREVKEGLHSSTPLHGSTGHHRQQ- 245
+ C H + +GFM ++ NY PS+ V+ TP + TG +
Sbjct 254 K-CAHHNNH-HEGFMNFMHRDEEI-NYYPSK----FDPVRCAEKVPTPTNSYTGIRTKCV 306
Query 246 -HPNSDFEQVGELYRRVMNDRQRDILITNIVSSLKH--ASREVQERQVKLFARADEDYGR 302
++F+Q G+ YR DRQ D + V L + E++ + +++AD G+
Sbjct 307 IKKENNFKQAGDRYRSWAPDRQ-DRFVKRWVEILSEPRLTHEIRGIWISYWSQADRSLGQ 365
Query 303 RVAKGLGLPLSV 314
++A L + S+
Sbjct 366 KLASRLNVRPSI 377
> eco:b1732 katE, ECK1730, JW1721; hydroperoxidase HPII(III) (catalase)
(EC:1.11.1.6); K03781 catalase [EC:1.11.1.6]
Length=753
Score = 194 bits (493), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 121/328 (36%), Positives = 178/328 (54%), Gaps = 25/328 (7%)
Query 1 VHQTTILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAER 60
+H SDRG P YR M G+ H F+ +N+E + + +FHWK G +L +EA++
Sbjct 250 LHNVMWAMSDRGIPRSYRTMEGFGIHTFRLINAEGKATFVRFHWKPLAGKASLVWDEAQK 309
Query 61 LAGTDPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRR 120
L G DPD+ R+L+ AIE GDFP + L Q++P + + +++LD TK+IP + P++R
Sbjct 310 LTGRDPDFHRRELWEAIEAGDFPEYELGFQLIPEEDEFKFDFDLLDPTKLIPEELVPVQR 369
Query 121 VGRLILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRL-GPNYD 179
VG+++LN NP N+F + EQ+AF P H++PG++ + D +LQGRLF+Y D RL GPN+
Sbjct 370 VGKMVLNRNPDNFFAENEQAAFHPGHIVPGLDFTNDPLLQGRLFSYTDTQISRLGGPNFH 429
Query 180 QIPINVPRGCPLHAFYIRDGF--MCVNGNYADLKNYTP-SEETNSLREVKEGLHSSTPLH 236
+IPIN P CP H F RDG M ++ N A NY P S N RE G P
Sbjct 430 EIPINRPT-CPYHNFQ-RDGMHRMGIDTNPA---NYEPNSINDNWPRETPPG-----PKR 479
Query 237 GSTGHHRQQHPNSDFEQ----VGELYR--RVMNDRQRDILITNIVSSLKHASREV----- 285
G ++++ + + GE Y R+ Q +IV +V
Sbjct 480 GGFESYQERVEGNKVRERSPSFGEYYSHPRLFWLSQTPFEQRHIVDGFSFELSKVVRPYI 539
Query 286 QERQVKLFARADEDYGRRVAKGLGLPLS 313
+ER V A D + VAK LG+ L+
Sbjct 540 RERVVDQLAHIDLTLAQAVAKNLGIELT 567
> ath:AT4G35090 CAT2; CAT2 (CATALASE 2); catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=474
Score = 186 bits (473), Expect = 8e-47, Method: Compositional matrix adjust.
Identities = 103/269 (38%), Positives = 157/269 (58%), Gaps = 11/269 (4%)
Query 5 TILFSDRGTPDGYRHMNGYSSHAFKPVNSENEVFYAKFHWKTNQGIKNLSSEEAERLAGT 64
T LF D G P YRHM+G + + +N + Y KFHWK G+K+L E+A R+ GT
Sbjct 187 TFLFDDIGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSLLEEDAIRVGGT 246
Query 65 DPDYATRDLFNAIERGDFPSWTLYIQVMPVREASHYRYNILDVTKVIPHKDYPLRRVGRL 124
+ +AT+DL+++I G++P W L+IQ++ + + ++ LDVTK P PL+ VGR+
Sbjct 247 NHSHATQDLYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKTWPEDILPLQPVGRM 306
Query 125 ILNVNPKNYFQDVEQSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQIPIN 184
+LN N N+F + EQ AF P+ ++PG+ S+DK+LQ R+F+Y D RHRLGPNY Q+P+N
Sbjct 307 VLNKNIDNFFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRLGPNYLQLPVN 366
Query 185 VPRGCPLHAFYIRDGFMCVNGNYADLK-NYTPSEETNSLREVKEGLHSSTPLHGSTGHHR 243
P+ C H + +GFM N + D + NY PS +V+ TP +G
Sbjct 367 APK-CAHHNNH-HEGFM--NFMHRDEEVNYFPSR----YDQVRHAEKYPTPPAVCSGKRE 418
Query 244 QQ--HPNSDFEQVGELYRRVMNDRQRDIL 270
+ ++F++ GE YR +RQ +
Sbjct 419 RCIIEKENNFKEPGERYRTFTPERQERFI 447
> ath:AT3G59630 diphthamide synthesis DPH2 family protein
Length=491
Score = 32.7 bits (73), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 272 TNIVSSLKHASREVQERQVKLFARADEDYGRRVAKGLG 309
T +VS+LK +R + +R+V+ F AD YG +G
Sbjct 39 TKVVSALKSKTRLLTDREVRFFVMADTTYGSCCIDEVG 76
> dre:436820 psmf1, zgc:92785; proteasome (prosome, macropain)
inhibitor subunit 1; K06700 proteasome inhibitor subunit 1
(PI31)
Length=272
Score = 31.2 bits (69), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 139 QSAFSPSHLIPGVEPSEDKMLQGRLFAYPDAHRHRLGPNYDQIP 182
+S F PS IPGV P R + RHR GP+ D +P
Sbjct 220 RSGFDPSSGIPGVLPPGAVPPGARFDPFGPVGRHRPGPDPDHMP 263
> cel:Y18D10A.1 hypothetical protein
Length=1634
Score = 30.8 bits (68), Expect = 6.2, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Query 218 ETNSLREVKEGLHSSTP-----LHGSTGHHRQQHPNSDFEQVGE 256
ET +++ EG+H T L ST HR Q P SDFE + +
Sbjct 329 ETKFVKKWPEGIHKPTEKDSFGLLNSTKIHRNQFPTSDFETIAQ 372
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13141051380
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40