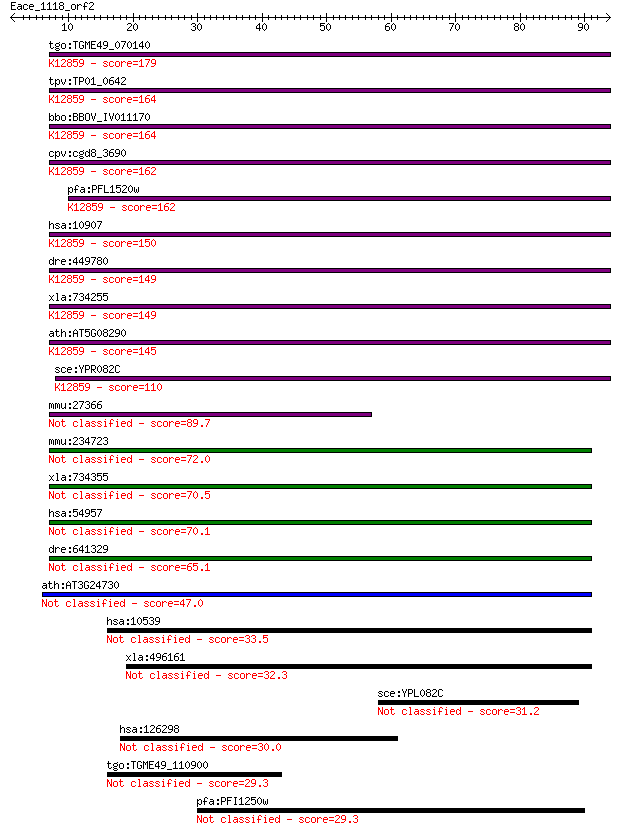
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1118_orf2
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070140 mitosis protein, putative ; K12859 U5 snRNP ... 179 2e-45
tpv:TP01_0642 mitosis protein Dim1; K12859 U5 snRNP protein, D... 164 5e-41
bbo:BBOV_IV011170 23.m06265; dim1 protein-like protein; K12859... 164 7e-41
cpv:cgd8_3690 mitosis protein DIM1 ; K12859 U5 snRNP protein, ... 162 2e-40
pfa:PFL1520w DIM1 protein homolog, putative; K12859 U5 snRNP p... 162 3e-40
hsa:10907 TXNL4A, DIB1, DIM1, HsT161, TXNL4, U5-15kD; thioredo... 150 1e-36
dre:449780 txnl4a, zgc:103632; thioredoxin-like 4A; K12859 U5 ... 149 2e-36
xla:734255 txnl4a, MGC85128; thioredoxin-like 4A; K12859 U5 sn... 149 2e-36
ath:AT5G08290 YLS8; YLS8; catalytic; K12859 U5 snRNP protein, ... 145 2e-35
sce:YPR082C DIB1, SNU16; 17-kDa component of the U4/U6aU5 tri-... 110 1e-24
mmu:27366 Txnl4a, D18Wsu98e, Dim1, Txnl4, U5-15kD, U5-15kDa; t... 89.7 2e-18
mmu:234723 Txnl4b, AI595343, D530025J19Rik, DLP, Dim2; thiored... 72.0 4e-13
xla:734355 txnl4b, MGC84953; thioredoxin-like 4B 70.5 1e-12
hsa:54957 TXNL4B, DLP, Dim2, FLJ20511; thioredoxin-like 4B 70.1 2e-12
dre:641329 MGC123253; zgc:123253 65.1 5e-11
ath:AT3G24730 catalytic 47.0 1e-05
hsa:10539 GLRX3, FLJ11864, GLRX4, GRX3, GRX4, PICOT, TXNL2, TX... 33.5 0.18
xla:496161 glrx3, txnl2; glutaredoxin 3 32.3
sce:YPL082C MOT1, BUR3, LPF4; Essential abundant protein invol... 31.2 0.82
hsa:126298 IRGQ, FLJ12521, FLJ38849, FLJ42796, FLJ46713, IRGQ1... 30.0 1.9
tgo:TGME49_110900 hypothetical protein 29.3 3.1
pfa:PFI1250w tlp2; Tlp2 29.3 3.5
> tgo:TGME49_070140 mitosis protein, putative ; K12859 U5 snRNP
protein, DIM1 family
Length=142
Score = 179 bits (455), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 83/87 (95%), Positives = 86/87 (98%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHD+DPECMKMDE L+KVAEDVKNFCVIY+
Sbjct 1 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDYDPECMKMDEQLYKVAEDVKNFCVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VDTTEVPDFTTMYELYDPVTVMFFYRN
Sbjct 61 VDTTEVPDFTTMYELYDPVTVMFFYRN 87
> tpv:TP01_0642 mitosis protein Dim1; K12859 U5 snRNP protein,
DIM1 family
Length=142
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 73/87 (83%), Positives = 81/87 (93%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYMLQHL SGW VDQAI+TEEER+VCIRFGHD DPEC+KMDE+LFK+AEDVKNFCVIY+
Sbjct 1 MSYMLQHLRSGWAVDQAIVTEEERVVCIRFGHDHDPECIKMDEILFKIAEDVKNFCVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF MYELYDP++VMFFYRN
Sbjct 61 VDITEVPDFNGMYELYDPISVMFFYRN 87
> bbo:BBOV_IV011170 23.m06265; dim1 protein-like protein; K12859
U5 snRNP protein, DIM1 family
Length=142
Score = 164 bits (415), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 73/87 (83%), Positives = 82/87 (94%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYMLQHL SGW VDQAI+TEEERLVCIRFGHD++PEC+KMDE+L+KVA+DVKNFCVIY+
Sbjct 1 MSYMLQHLRSGWAVDQAIVTEEERLVCIRFGHDYNPECIKMDEVLYKVADDVKNFCVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF MYELYDPV+VMFFYRN
Sbjct 61 VDITEVPDFNGMYELYDPVSVMFFYRN 87
> cpv:cgd8_3690 mitosis protein DIM1 ; K12859 U5 snRNP protein,
DIM1 family
Length=142
Score = 162 bits (411), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 72/87 (82%), Positives = 82/87 (94%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYMLQHLH+GW VDQAIL+EEER+VCIRFGHD+D +CM MDE+L+KVAEDVKNFCVIY+
Sbjct 1 MSYMLQHLHNGWSVDQAILSEEERIVCIRFGHDYDADCMLMDEVLYKVAEDVKNFCVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF TMYELYDPV+VMFF+RN
Sbjct 61 VDITEVPDFNTMYELYDPVSVMFFFRN 87
> pfa:PFL1520w DIM1 protein homolog, putative; K12859 U5 snRNP
protein, DIM1 family
Length=139
Score = 162 bits (409), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 73/84 (86%), Positives = 79/84 (94%), Gaps = 0/84 (0%)
Query 10 MLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVVDT 69
ML HLHSGW VDQAIL+EEERLVCIRFGHD+DP+CMKMDELLFKV ED+KNFCVIY+VD
Sbjct 1 MLYHLHSGWDVDQAILSEEERLVCIRFGHDYDPDCMKMDELLFKVVEDIKNFCVIYLVDI 60
Query 70 TEVPDFTTMYELYDPVTVMFFYRN 93
TEVPDF TMYELYDPV+VMFFYRN
Sbjct 61 TEVPDFNTMYELYDPVSVMFFYRN 84
> hsa:10907 TXNL4A, DIB1, DIM1, HsT161, TXNL4, U5-15kD; thioredoxin-like
4A; K12859 U5 snRNP protein, DIM1 family
Length=142
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 67/87 (77%), Positives = 77/87 (88%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYML HLH+GWQVDQAIL+EE+R+V IRFGHD+DP CMKMDE+L+ +AE VKNF VIY+
Sbjct 1 MSYMLPHLHNGWQVDQAILSEEDRVVVIRFGHDWDPTCMKMDEVLYSIAEKVKNFAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF MYELYDP TVMFF+RN
Sbjct 61 VDITEVPDFNKMYELYDPCTVMFFFRN 87
> dre:449780 txnl4a, zgc:103632; thioredoxin-like 4A; K12859 U5
snRNP protein, DIM1 family
Length=142
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 66/87 (75%), Positives = 76/87 (87%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYML HLH+GWQVDQAIL+EE+R + IRFGHD+DP CMKMDE+L+ +AE VKNF VIY+
Sbjct 1 MSYMLPHLHNGWQVDQAILSEEDRAIVIRFGHDWDPTCMKMDEVLYSIAEKVKNFAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF MYELYDP TVMFF+RN
Sbjct 61 VDITEVPDFNKMYELYDPCTVMFFFRN 87
> xla:734255 txnl4a, MGC85128; thioredoxin-like 4A; K12859 U5
snRNP protein, DIM1 family
Length=142
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 66/87 (75%), Positives = 77/87 (88%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSYML HLH+GWQVDQAIL+EE+R++ IRFGHD+DP CMKMDE+L+ +AE VKNF VIY+
Sbjct 1 MSYMLPHLHNGWQVDQAILSEEDRVLVIRFGHDWDPTCMKMDEVLYSIAEKVKNFAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF MYELYDP TVMFF+RN
Sbjct 61 VDITEVPDFNKMYELYDPCTVMFFFRN 87
> ath:AT5G08290 YLS8; YLS8; catalytic; K12859 U5 snRNP protein,
DIM1 family
Length=142
Score = 145 bits (367), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 66/87 (75%), Positives = 74/87 (85%), Gaps = 0/87 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MSY+L HLHSGW VDQ+IL EEERLV IRFGHD+D CM+MDE+L VAE +KNF VIY+
Sbjct 1 MSYLLPHLHSGWAVDQSILAEEERLVVIRFGHDWDETCMQMDEVLASVAETIKNFAVIYL 60
Query 67 VDTTEVPDFTTMYELYDPVTVMFFYRN 93
VD TEVPDF TMYELYDP TVMFF+RN
Sbjct 61 VDITEVPDFNTMYELYDPSTVMFFFRN 87
> sce:YPR082C DIB1, SNU16; 17-kDa component of the U4/U6aU5 tri-snRNP,
plays an essential role in pre-mRNA splicing, orthologue
of hDIM1, the human U5-specific 15-kDa protein; K12859
U5 snRNP protein, DIM1 family
Length=143
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 53/86 (61%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 8 SYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVV 67
S +L L +GW VDQAI+TE +RLV IRFG D +CM MDELL +AE V+NF VIY+
Sbjct 3 SVLLPQLRTGWHVDQAIVTETKRLVVIRFGRKNDRQCMIMDELLSSIAERVRNFAVIYLC 62
Query 68 DTTEVPDFTTMYELYDPVTVMFFYRN 93
D EV DF MYEL DP+TVMFFY N
Sbjct 63 DIDEVSDFDEMYELTDPMTVMFFYHN 88
> mmu:27366 Txnl4a, D18Wsu98e, Dim1, Txnl4, U5-15kD, U5-15kDa;
thioredoxin-like 4A
Length=63
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 38/50 (76%), Positives = 46/50 (92%), Gaps = 0/50 (0%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAE 56
MSYML HLH+GWQVDQAIL+EE+R+V IRFGHD+DP CMKMDE+L+ +AE
Sbjct 1 MSYMLPHLHNGWQVDQAILSEEDRVVVIRFGHDWDPTCMKMDEVLYSIAE 50
> mmu:234723 Txnl4b, AI595343, D530025J19Rik, DLP, Dim2; thioredoxin-like
4B
Length=149
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 54/85 (63%), Gaps = 1/85 (1%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MS++L L S +VDQAI + E+++ +RFG D DP C+++D++L K + D+ IY+
Sbjct 1 MSFLLPKLTSKKEVDQAIKSTAEKVLVLRFGRDEDPVCLQLDDILSKTSADLSKMAAIYL 60
Query 67 VDTTEVPDFTTMYEL-YDPVTVMFF 90
VD P +T +++ Y P TV FF
Sbjct 61 VDVDHTPVYTQYFDISYIPSTVFFF 85
> xla:734355 txnl4b, MGC84953; thioredoxin-like 4B
Length=149
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 54/85 (63%), Gaps = 1/85 (1%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MS++L L S VDQAI T E+++ +RFG D D C+++D++L K + D+ IY+
Sbjct 1 MSFLLPKLSSKRDVDQAIKTTAEKVLVLRFGRDEDHVCLQLDDILSKTSHDLSKMASIYI 60
Query 67 VDTTEVPDFTTMYEL-YDPVTVMFF 90
VD +VP +T +++ Y P T+ FF
Sbjct 61 VDVDKVPVYTQYFDISYIPSTIFFF 85
> hsa:54957 TXNL4B, DLP, Dim2, FLJ20511; thioredoxin-like 4B
Length=149
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 54/85 (63%), Gaps = 1/85 (1%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MS++L L S +VDQAI + E+++ +RFG D DP C+++D++L K + D+ IY+
Sbjct 1 MSFLLPKLTSKKEVDQAIKSTAEKVLVLRFGRDEDPVCLQLDDILSKTSSDLSKMAAIYL 60
Query 67 VDTTEVPDFTTMYEL-YDPVTVMFF 90
VD + +T +++ Y P TV FF
Sbjct 61 VDVDQTAVYTQYFDISYIPSTVFFF 85
> dre:641329 MGC123253; zgc:123253
Length=149
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 51/85 (60%), Gaps = 1/85 (1%)
Query 7 MSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV 66
MS L L S +D+AI + E+++ +RFG D D C+++DE+L K A D+ N IY+
Sbjct 1 MSLFLPKLSSKKDIDEAIKSVAEKVLVLRFGRDEDSVCLQIDEILSKTAHDLSNMASIYL 60
Query 67 VDTTEVPDFTTMYEL-YDPVTVMFF 90
VD P ++ +++ + P TV FF
Sbjct 61 VDVDSAPVYSRYFDISFIPSTVFFF 85
> ath:AT3G24730 catalytic
Length=159
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 47/89 (52%), Gaps = 5/89 (5%)
Query 6 KMSYMLQHLHSGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIY 65
+MSY+L+ L + ++D+ I + ++ +RFG D C++ DE+L K DV F +
Sbjct 8 EMSYLLKTLTTKEEIDRVIRDTIDEVLVLRFGRSSDAVCLQHDEILAKSVRDVSKFAKVA 67
Query 66 VVDTTEVPDFTTMYELYD----PVTVMFF 90
+VD + D + +D P T+ FF
Sbjct 68 LVD-VDSEDVQVYVKYFDITLFPSTIFFF 95
> hsa:10539 GLRX3, FLJ11864, GLRX4, GRX3, GRX4, PICOT, TXNL2,
TXNL3; glutaredoxin 3
Length=335
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 16/75 (21%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Query 16 SGWQVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVVDTTEVPDF 75
S Q ++ + + + L+ + F + P+C +M+E++ ++A+++ + ++ VP+
Sbjct 18 SAGQFEELLRLKAKSLLVVHFWAPWAPQCAQMNEVMAELAKELPQVSFVK-LEAEGVPEV 76
Query 76 TTMYELYDPVTVMFF 90
+ YE+ T +FF
Sbjct 77 SEKYEISSVPTFLFF 91
> xla:496161 glrx3, txnl2; glutaredoxin 3
Length=326
Score = 32.3 bits (72), Expect = 0.41, Method: Composition-based stats.
Identities = 17/73 (23%), Positives = 40/73 (54%), Gaps = 3/73 (4%)
Query 19 QVDQAILTEEERLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYV-VDTTEVPDFTT 77
Q ++ + + L + F + P+C +M+E++ ++A++ V++V ++ VP+ +
Sbjct 12 QFEELLQKSAKSLTVVHFWAPWAPQCTQMNEVMAELAKEQPQ--VMFVKLEAEAVPEVSE 69
Query 78 MYELYDPVTVMFF 90
YE+ T +FF
Sbjct 70 KYEITSVPTFLFF 82
> sce:YPL082C MOT1, BUR3, LPF4; Essential abundant protein involved
in regulation of transcription, removes Spt15p (TBP) from
DNA via its C-terminal ATPase activity, forms a complex
with TBP that binds TATA DNA with high affinity but with altered
specificity (EC:3.6.1.-)
Length=1867
Score = 31.2 bits (69), Expect = 0.82, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 58 VKNFCVIYVVDTTEVPDFTTMYELYDPVTVM 88
VKN C VDT+EVPDF+ E + + +
Sbjct 979 VKNLCGFLCVDTSEVPDFSVNAEYKEKILTL 1009
> hsa:126298 IRGQ, FLJ12521, FLJ38849, FLJ42796, FLJ46713, IRGQ1;
immunity-related GTPase family, Q
Length=623
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 18 WQVDQAILTEEERLVCIRF-GHDFDPECMKMDELLFKVAEDVKN 60
W QA+L + LVC+R G DPEC+ ++ E +KN
Sbjct 313 WAQVQALLLPDAPLVCVRTDGEGEDPECLGEGKMENPKGESLKN 356
> tgo:TGME49_110900 hypothetical protein
Length=2243
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 16 SGWQVDQAILTEEERLVCIRFGHDFDP 42
S W+ + A T EER + RFGH P
Sbjct 1165 SEWRKETASFTSEERALIARFGHLLSP 1191
> pfa:PFI1250w tlp2; Tlp2
Length=128
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 30 RLVCIRFGHDFDPECMKMDELLFKVAEDVKNFCVIYVVDTTEVPDFTTMYELYDPVTVMF 89
+LV +FG + C KM ++ K+ ED N +Y +D E P+ ++ + T++
Sbjct 42 KLVVAQFGASWCAPCKKMKPVIEKLGEDNDNIESLY-IDIDEFPELGENEDINELPTILL 100
Lambda K H
0.326 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2074765676
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40