bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1141_orf1
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
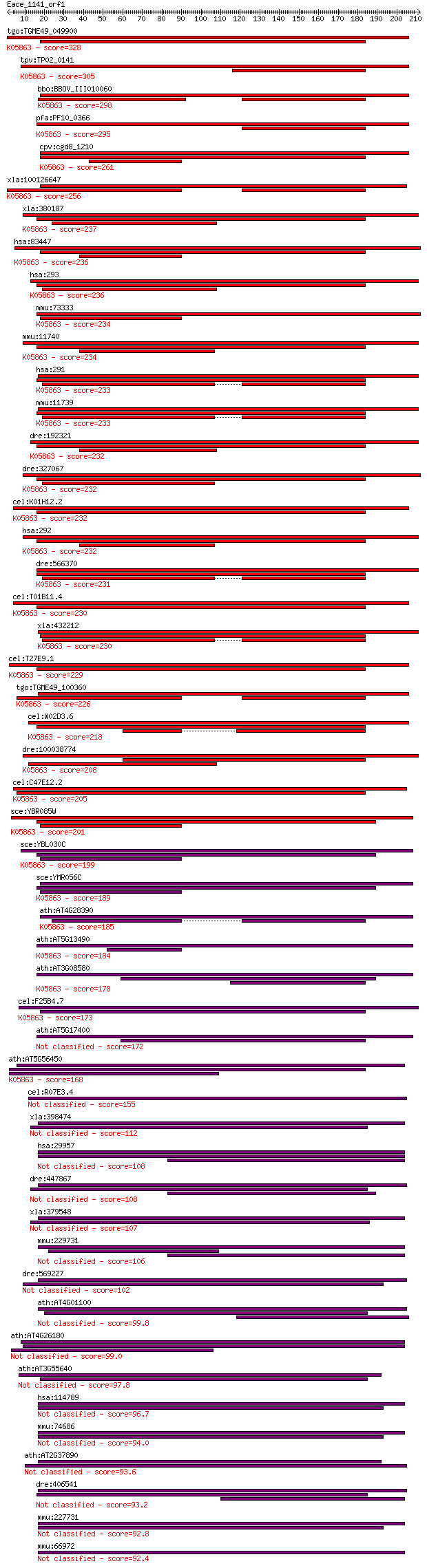
tgo:TGME49_049900 ADP/ATP carrier, putative ; K05863 solute ca... 328 1e-89
tpv:TP02_0141 adenine nucleotide translocase; K05863 solute ca... 305 9e-83
bbo:BBOV_III010060 17.m07874; adenine nucleotide translocase; ... 298 1e-80
pfa:PF10_0366 ADP/ATP transporter on adenylate translocase; K0... 295 6e-80
cpv:cgd8_1210 mitochondrial ADP/ATP-transporter, integral memb... 261 1e-69
xla:100126647 hypothetical protein LOC100126647; K05863 solute... 256 4e-68
xla:380187 slc25a5, MGC53201; solute carrier family 25 (mitoch... 237 2e-62
hsa:83447 SLC25A31, AAC4, ANT4, DKFZp434N1235, SFEC35kDa; solu... 236 3e-62
hsa:293 SLC25A6, AAC3, ANT3, ANT3Y, MGC17525; solute carrier f... 236 5e-62
mmu:73333 Slc25a31, 1700034J06Rik, Ant4, Sfec; solute carrier ... 234 2e-61
mmu:11740 Slc25a5, Ant2, MGC101927; solute carrier family 25 (... 234 2e-61
hsa:291 SLC25A4, AAC1, ANT, ANT1, PEO2, PEO3, T1; solute carri... 233 3e-61
mmu:11739 Slc25a4, AU019225, Ant1; solute carrier family 25 (m... 233 3e-61
dre:192321 slc25a5, CHUNP6931, SI:bZ46J2.1, SI:bZ46J2.3, SI:bZ... 232 6e-61
dre:327067 slc25a4, fa22e07, wu:fa22e07, zgc:77591; solute car... 232 6e-61
cel:K01H12.2 ant-1.3; Adenine Nucleotide Translocator family m... 232 6e-61
hsa:292 SLC25A5, 2F1, AAC2, ANT2, T2, T3; solute carrier famil... 232 7e-61
dre:566370 slc25a6, si:dkey-21o13.4, wu:fj78b08; solute carrie... 231 1e-60
cel:T01B11.4 ant-1.44; Adenine Nucleotide Translocator family ... 230 2e-60
xla:432212 slc25a4, MGC79005; solute carrier family 25 (mitoch... 230 2e-60
cel:T27E9.1 ant-1.1; Adenine Nucleotide Translocator family me... 229 4e-60
tgo:TGME49_100360 mitochondrial carrier domain-containing prot... 226 5e-59
cel:W02D3.6 ant-1.2; Adenine Nucleotide Translocator family me... 218 9e-57
dre:100038774 si:dkey-251i10.1; K05863 solute carrier family 2... 208 8e-54
cel:C47E12.2 hypothetical protein; K05863 solute carrier famil... 205 8e-53
sce:YBR085W AAC3, ANC3; Aac3p; K05863 solute carrier family 25... 201 1e-51
sce:YBL030C PET9, AAC2, ANC2; Major ADP/ATP carrier of the mit... 199 6e-51
sce:YMR056C AAC1; Aac1p; K05863 solute carrier family 25 (mito... 189 5e-48
ath:AT4G28390 AAC3; AAC3 (ADP/ATP CARRIER 3); ATP:ADP antiport... 185 1e-46
ath:AT5G13490 AAC2; AAC2 (ADP/ATP carrier 2); ATP:ADP antiport... 184 2e-46
ath:AT3G08580 AAC1; AAC1 (ADP/ATP CARRIER 1); ATP:ADP antiport... 178 1e-44
cel:F25B4.7 hypothetical protein; K05863 solute carrier family... 173 5e-43
ath:AT5G17400 ER-ANT1 (ENDOPLASMIC RETICULUM-DENINE NUCLEOTIDE... 172 1e-42
ath:AT5G56450 mitochondrial substrate carrier family protein; ... 168 1e-41
cel:R07E3.4 hypothetical protein 155 7e-38
xla:398474 slc25a24-a, MGC132369, apc1, scamc-1, scamc1-A, slc... 112 1e-24
hsa:29957 SLC25A24, APC1, DKFZp586G0123, SCAMC-1; solute carri... 108 1e-23
dre:447867 slc25a24; zgc:92470; K14684 solute carrier family 2... 108 2e-23
xla:379548 slc25a24-b, MGC68982, apc1, scamc-1, scamc1-B; solu... 107 3e-23
mmu:229731 Slc25a24, 2610016M12Rik; solute carrier family 25 (... 106 5e-23
dre:569227 slc25a25b, scamc2b, si:dkey-7o20.1; solute carrier ... 102 7e-22
ath:AT4G01100 ADNT1; ADNT1 (ADENINE NUCLEOTIDE TRANSPORTER 1);... 99.8 6e-21
ath:AT4G26180 mitochondrial substrate carrier family protein 99.0 1e-20
ath:AT3G55640 mitochondrial substrate carrier family protein 97.8 3e-20
hsa:114789 SLC25A25, KIAA1896, MCSC, MGC105138, MGC119514, MGC... 96.7 6e-20
mmu:74686 4930443G12Rik; RIKEN cDNA 4930443G12 gene; K14684 so... 94.0 4e-19
ath:AT2G37890 mitochondrial substrate carrier family protein 93.6 4e-19
dre:406541 slc25a25a, slc25a, slc25a25, wu:fb13d12, wu:fd14e03... 93.2 6e-19
mmu:227731 Slc25a25, 1110030N17Rik, MCSC, MGC36388, mKIAA1896;... 92.8 7e-19
mmu:66972 Slc25a23, 2310067G05Rik; solute carrier family 25 (m... 92.4 1e-18
> tgo:TGME49_049900 ADP/ATP carrier, putative ; K05863 solute
carrier family 25 (mitochondrial adenine nucleotide translocator),
member 4/5/6/31
Length=318
Score = 328 bits (840), Expect = 1e-89, Method: Compositional matrix adjust.
Identities = 154/205 (75%), Positives = 181/205 (88%), Gaps = 0/205 (0%)
Query 1 GKEEKRRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRY 60
GKE+K ++ L++ AK+F+AGG+SAGVSKTIVAP+ERVKMLIQTQDSIP I++G PRY
Sbjct 10 GKEKKGNADSQLMNTAKDFLAGGISAGVSKTIVAPIERVKMLIQTQDSIPEIKEGKIPRY 69
Query 61 SGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKF 120
+GI++CF RV EQGV SLWRGN+ANVIRYFPTQAFNF FKDTFK++FP+Y+Q +EF+KF
Sbjct 70 TGIVDCFRRVSAEQGVASLWRGNMANVIRYFPTQAFNFAFKDTFKRMFPRYDQKKEFWKF 129
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSL 180
F TNV SGGLAGA+SL +VYPLDFARTRLASDVGKG+ REF GL DC+ K+ RRTGF SL
Sbjct 130 FCTNVASGGLAGASSLVIVYPLDFARTRLASDVGKGNEREFTGLVDCLGKIFRRTGFFSL 189
Query 181 YQGFGVSVQGIIVYRGAYFGLYDAA 205
YQGFGVSVQGIIVYRGAYFGL+D A
Sbjct 190 YQGFGVSVQGIIVYRGAYFGLFDTA 214
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 40/169 (23%), Positives = 74/169 (43%), Gaps = 12/169 (7%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
N +GG++ S IV P++ + + + + G+ ++G+++C ++ + G +
Sbjct 133 NVASGGLAGASSLVIVYPLDFARTRLASD-----VGKGNEREFTGLVDCLGKIFRRTGFF 187
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
SL++G +V + F DT K + N + F + AG AS
Sbjct 188 SLYQGFGVSVQGIIVYRGAYFGLFDTAKAMLFGPNNDANLFYKWAVAQTVTAAAGVAS-- 245
Query 138 VVYPLDFARTRLASDVGKGSAR---EFLGLGDCIKKVSRRTGFLSLYQG 183
YP D R R+ G+ ++ G DC KKV ++ GF ++G
Sbjct 246 --YPFDTVRRRMMMMAGRKKGEQEIQYTGTADCWKKVYQQEGFKGFFKG 292
> tpv:TP02_0141 adenine nucleotide translocase; K05863 solute
carrier family 25 (mitochondrial adenine nucleotide translocator),
member 4/5/6/31
Length=301
Score = 305 bits (780), Expect = 9e-83, Method: Compositional matrix adjust.
Identities = 145/198 (73%), Positives = 167/198 (84%), Gaps = 4/198 (2%)
Query 8 DNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCF 67
+ N L+D F+ GGVSA +SKT VAP+ERVKMLIQTQDSIP I+ G PRYSGI+NCF
Sbjct 4 NKNFLVD----FLMGGVSAAISKTAVAPIERVKMLIQTQDSIPDIKSGKVPRYSGILNCF 59
Query 68 SRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLS 127
+RV KEQGV SLWRGNLANVIRYFPTQAFNF FKD FK++FPKYNQ +F+KFF N+ S
Sbjct 60 ARVSKEQGVTSLWRGNLANVIRYFPTQAFNFAFKDYFKRMFPKYNQKTDFWKFFGANLAS 119
Query 128 GGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVS 187
GGLAGA+SL +VYPLDFARTRLASDVGKG+ REF GL DC+ K+ R TG +SLY+GF VS
Sbjct 120 GGLAGASSLLIVYPLDFARTRLASDVGKGAKREFTGLLDCLMKIQRSTGVMSLYKGFMVS 179
Query 188 VQGIIVYRGAYFGLYDAA 205
VQGIIVYRGAYFG+YD+A
Sbjct 180 VQGIIVYRGAYFGMYDSA 197
Score = 38.1 bits (87), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 38/73 (52%), Gaps = 5/73 (6%)
Query 116 EFFKFFLTNVLSGGLAGAASLTVVYPLDFARTRLAS-----DVGKGSAREFLGLGDCIKK 170
E K FL + L GG++ A S T V P++ + + + D+ G + G+ +C +
Sbjct 2 EVNKNFLVDFLMGGVSAAISKTAVAPIERVKMLIQTQDSIPDIKSGKVPRYSGILNCFAR 61
Query 171 VSRRTGFLSLYQG 183
VS+ G SL++G
Sbjct 62 VSKEQGVTSLWRG 74
> bbo:BBOV_III010060 17.m07874; adenine nucleotide translocase;
K05863 solute carrier family 25 (mitochondrial adenine nucleotide
translocator), member 4/5/6/31
Length=300
Score = 298 bits (762), Expect = 1e-80, Method: Compositional matrix adjust.
Identities = 138/188 (73%), Positives = 157/188 (83%), Gaps = 0/188 (0%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+F+ GGVSA VSKT VAP+ERVKMLIQTQD+IP I+ G PRY+GI+NCF RVC EQGV
Sbjct 10 DFLMGGVSAAVSKTAVAPIERVKMLIQTQDTIPEIKSGKLPRYTGIVNCFGRVCAEQGVS 69
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
SLWRGN+ANVIRYFPTQAFNF FKD FK +FPKYNQ EF+KFF NV SGGLAGA+SL
Sbjct 70 SLWRGNMANVIRYFPTQAFNFAFKDFFKTLFPKYNQKTEFWKFFAANVASGGLAGASSLM 129
Query 138 VVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRGA 197
+VYPLDFARTRLASDV K REF GL DC+ K+ R TGF+SLY+GF +SV GIIVYRG
Sbjct 130 IVYPLDFARTRLASDVRKEGQREFTGLLDCLMKIKRSTGFMSLYKGFSISVTGIIVYRGT 189
Query 198 YFGLYDAA 205
YFG+YD+A
Sbjct 190 YFGMYDSA 197
Score = 33.9 bits (76), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 8/77 (10%)
Query 17 KNFIAGGVS--AGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQ 74
K F+A V+ AG++ V R M++ + I Y+ ++CF ++ K +
Sbjct 213 KWFVAQTVTINAGLASYPFDTVRRRMMMMSGKKQTSEIM------YTSSLDCFRKILKNE 266
Query 75 GVYSLWRGNLANVIRYF 91
GV ++G AN++R F
Sbjct 267 GVNGFFKGAFANILRGF 283
Score = 33.1 bits (74), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 5/68 (7%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLAS-----DVGKGSAREFLGLGDCIKKVSRRT 175
F+T+ L GG++ A S T V P++ + + + ++ G + G+ +C +V
Sbjct 7 FVTDFLMGGVSAAVSKTAVAPIERVKMLIQTQDTIPEIKSGKLPRYTGIVNCFGRVCAEQ 66
Query 176 GFLSLYQG 183
G SL++G
Sbjct 67 GVSSLWRG 74
> pfa:PF10_0366 ADP/ATP transporter on adenylate translocase;
K05863 solute carrier family 25 (mitochondrial adenine nucleotide
translocator), member 4/5/6/31
Length=301
Score = 295 bits (756), Expect = 6e-80, Method: Compositional matrix adjust.
Identities = 137/190 (72%), Positives = 159/190 (83%), Gaps = 0/190 (0%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A +F+ GG+SA +SKT+VAP+ERVKMLIQTQDSIP I+ G RYSG+INCF RV KEQG
Sbjct 10 AADFLMGGISAAISKTVVAPIERVKMLIQTQDSIPEIKSGQVERYSGLINCFKRVSKEQG 69
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
V SLWRGN+ANVIRYFPTQAFNF FKD FK IFP+Y+QN +F KFF N+LSG AGA S
Sbjct 70 VLSLWRGNVANVIRYFPTQAFNFAFKDYFKNIFPRYDQNTDFSKFFCVNILSGATAGAIS 129
Query 136 LTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYR 195
L +VYPLDFARTRLASD+GKG R+F GL DC+ K+ ++TG LSLY GFGVSV GIIVYR
Sbjct 130 LLIVYPLDFARTRLASDIGKGKDRQFTGLFDCLAKIYKQTGLLSLYSGFGVSVTGIIVYR 189
Query 196 GAYFGLYDAA 205
G+YFGLYD+A
Sbjct 190 GSYFGLYDSA 199
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 5/68 (7%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLAS-----DVGKGSAREFLGLGDCIKKVSRRT 175
F + L GG++ A S TVV P++ + + + ++ G + GL +C K+VS+
Sbjct 9 FAADFLMGGISAAISKTVVAPIERVKMLIQTQDSIPEIKSGQVERYSGLINCFKRVSKEQ 68
Query 176 GFLSLYQG 183
G LSL++G
Sbjct 69 GVLSLWRG 76
> cpv:cgd8_1210 mitochondrial ADP/ATP-transporter, integral membrane
protein with 4 transmembrane domains ; K05863 solute
carrier family 25 (mitochondrial adenine nucleotide translocator),
member 4/5/6/31
Length=325
Score = 261 bits (666), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 120/188 (63%), Positives = 148/188 (78%), Gaps = 0/188 (0%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+FI GG+SA +SKT VAP+ERVK+L+QTQD+ P I G PRY+GI +C RV KEQG+
Sbjct 35 DFIVGGISATISKTAVAPIERVKLLLQTQDTNPDIIKGLIPRYAGIFDCLRRVSKEQGIL 94
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
SLWRGN NVIRYFPTQAF F FKD + + P+YN+ +F+KFF N+LSGGLAGAAS
Sbjct 95 SLWRGNTTNVIRYFPTQAFGFAFKDMIRDMMPRYNKESDFWKFFGVNMLSGGLAGAASSG 154
Query 138 VVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRGA 197
+VYPLDFARTRLA+D+GK ++EF G+ DCI K+S+++G SLYQGF VS+QGI VYR A
Sbjct 155 IVYPLDFARTRLATDIGKNGSKEFKGMFDCIMKISKQSGIRSLYQGFFVSIQGIFVYRAA 214
Query 198 YFGLYDAA 205
YFGLYD
Sbjct 215 YFGLYDTT 222
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 45/169 (26%), Positives = 85/169 (50%), Gaps = 14/169 (8%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
N ++GG++ S IV P++ + + T I + + G+ +C ++ K+ G+
Sbjct 141 NMLSGGLAGAASSGIVYPLDFARTRLATD-----IGKNGSKEFKGMFDCIMKISKQSGIR 195
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEE--FFKFFLTNVLSGGLAGAAS 135
SL++G ++ F +A F DT K++F K NQ +E +K+ + ++ ++
Sbjct 196 SLYQGFFVSIQGIFVYRAAYFGLYDTTKEMFFK-NQKQENMLYKWIIAQTVT-----TSA 249
Query 136 LTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQG 183
+ YP D R R+ GK G + G DC+KK+ R+ G +L++G
Sbjct 250 GIICYPFDTIRRRMMMMAGKKGKDVLYTGAYDCLKKIIRKEGVGALFKG 298
Score = 35.0 bits (79), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 43 IQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIR 89
I+ + + A + G Y+G +C ++ +++GV +L++G+L+NV+R
Sbjct 259 IRRRMMMMAGKKGKDVLYTGAYDCLKKIIRKEGVGALFKGSLSNVLR 305
> xla:100126647 hypothetical protein LOC100126647; K05863 solute
carrier family 25 (mitochondrial adenine nucleotide translocator),
member 4/5/6/31
Length=306
Score = 256 bits (654), Expect = 4e-68, Method: Compositional matrix adjust.
Identities = 125/188 (66%), Positives = 146/188 (77%), Gaps = 2/188 (1%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+F+ GGVS ++KT AP+ERVK++IQTQDS P I G PRY+GI NCF+RV +EQG
Sbjct 13 DFLLGGVSGAIAKTATAPIERVKLIIQTQDSNPRIISGEVPRYTGIGNCFTRVYQEQGFA 72
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
+ WRGN N+IRYFPTQAFNF FKDT KK+FPK N EEF KFFL N+ SGGLAGA SL
Sbjct 73 AFWRGNFTNIIRYFPTQAFNFAFKDTIKKLFPKVNPKEEFGKFFLVNMASGGLAGAGSLC 132
Query 138 VVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSR-RTGFLSLYQGFGVSVQGIIVYRG 196
+VYPLD+ARTRLASDVG G AR+F GLGDC+ K +R G L LY GFGVSV GII YRG
Sbjct 133 IVYPLDYARTRLASDVGSG-ARDFNGLGDCLVKTARGPKGILGLYNGFGVSVAGIIPYRG 191
Query 197 AYFGLYDA 204
YFG+YD+
Sbjct 192 VYFGMYDS 199
Score = 38.1 bits (87), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 42/95 (44%), Gaps = 10/95 (10%)
Query 1 GKEEKRRDNNP------LIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRD 54
G + R+ NP +I A F +A + P + ++ +Q Q P
Sbjct 195 GMYDSLREKNPYKKDTGIIGLASKFAVAQFTAICAGYASYPFDTIRRRLQMQSEKPK--- 251
Query 55 GHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIR 89
Y G ++CF ++ K +GV ++++G AN +R
Sbjct 252 -EQWLYKGTVDCFGKIMKNEGVTAMFKGAGANALR 285
Score = 35.0 bits (79), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 36/68 (52%), Gaps = 5/68 (7%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASD-----VGKGSAREFLGLGDCIKKVSRRT 175
F+T+ L GG++GA + T P++ + + + + G + G+G+C +V +
Sbjct 10 FVTDFLLGGVSGAIAKTATAPIERVKLIIQTQDSNPRIISGEVPRYTGIGNCFTRVYQEQ 69
Query 176 GFLSLYQG 183
GF + ++G
Sbjct 70 GFAAFWRG 77
> xla:380187 slc25a5, MGC53201; solute carrier family 25 (mitochondrial
carrier; adenine nucleotide translocator), member
5; K05863 solute carrier family 25 (mitochondrial adenine nucleotide
translocator), member 4/5/6/31
Length=298
Score = 237 bits (604), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 122/208 (58%), Positives = 150/208 (72%), Gaps = 9/208 (4%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIR-DGHTPRYSGIINCF 67
+ I AK+F+AGGV+A +SKT VAP+ERVK+L+Q Q + I D H Y GI++C
Sbjct 2 TDAAISFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITADKH---YKGIMDCV 58
Query 68 SRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVL 126
R+ KEQG S WRGNLANVIRYFPTQA NF FKD +KKIF ++ +F+++F N+
Sbjct 59 VRIPKEQGFVSFWRGNLANVIRYFPTQALNFAFKDKYKKIFLDNVDKKTQFWRYFAGNLA 118
Query 127 SGGLAGAASLTVVYPLDFARTRLASDVGKGS-AREFLGLGDCIKKVSRRTGFLSLYQGFG 185
SGG AGA SL VYPLDFARTRLA+DVGKG+ REF GLGDC+ K+S+ G LYQGF
Sbjct 119 SGGAAGATSLCFVYPLDFARTRLAADVGKGANEREFKGLGDCLVKISKSDGIKGLYQGFN 178
Query 186 VSVQGIIVYRGAYFGLYDAA---VPAPN 210
VSVQGII+YR AYFG+YD A +P P
Sbjct 179 VSVQGIIIYRAAYFGIYDTAKGMLPDPK 206
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 75/169 (44%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + + + G+ +C ++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKGANEREFKGLGDCLVKISKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P F + + ++ +AG AS
Sbjct 170 IKGLYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKNTHIFVSWMIAQTVTA-VAGFAS 228
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
YP D R R+ G KG+ + G DC KK++R G + ++G
Sbjct 229 ----YPFDTVRRRMMMQSGRKGADIMYSGTIDCWKKIARDEGSKAFFKG 273
Score = 37.4 bits (85), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 24/91 (26%), Positives = 44/91 (48%), Gaps = 12/91 (13%)
Query 24 VSAGVSKTIVA-------PVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
VS +++T+ A P + V+ + Q R G YSG I+C+ ++ +++G
Sbjct 212 VSWMIAQTVTAVAGFASYPFDTVRRRMMMQSG----RKGADIMYSGTIDCWKKIARDEGS 267
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKI 107
+ ++G +NV+R AF D KK+
Sbjct 268 KAFFKGAWSNVLRGMGG-AFVLVLYDELKKV 297
> hsa:83447 SLC25A31, AAC4, ANT4, DKFZp434N1235, SFEC35kDa; solute
carrier family 25 (mitochondrial carrier; adenine nucleotide
translocator), member 31; K05863 solute carrier family
25 (mitochondrial adenine nucleotide translocator), member
4/5/6/31
Length=315
Score = 236 bits (603), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 120/215 (55%), Positives = 153/215 (71%), Gaps = 10/215 (4%)
Query 5 KRRDNNPLIDAA---KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYS 61
K++ L DA+ K+ +AGGV+A VSKT VAP+ERVK+L+Q Q S I RY
Sbjct 7 KKKAEKRLFDASSFGKDLLAGGVAAAVSKTAVAPIERVKLLLQVQASSKQISP--EARYK 64
Query 62 GIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFKF 120
G+++C R+ +EQG +S WRGNLANVIRYFPTQA NF FKD +K++F N+ ++F+++
Sbjct 65 GMVDCLVRIPREQGFFSFWRGNLANVIRYFPTQALNFAFKDKYKQLFMSGVNKEKQFWRW 124
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKG-SAREFLGLGDCIKKVSRRTGFLS 179
FL N+ SGG AGA SL VVYPLDFARTRL D+GKG R+F GLGDCI K+++ G
Sbjct 125 FLANLASGGAAGATSLCVVYPLDFARTRLGVDIGKGPEERQFKGLGDCIMKIAKSDGIAG 184
Query 180 LYQGFGVSVQGIIVYRGAYFGLYDAA---VPAPNQ 211
LYQGFGVSVQGIIVYR +YFG YD +P P +
Sbjct 185 LYQGFGVSVQGIIVYRASYFGAYDTVKGLLPKPKK 219
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/166 (23%), Positives = 74/166 (44%), Gaps = 10/166 (6%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
N +GG + S +V P++ +T+ + + ++ G+ +C ++ K G+
Sbjct 128 NLASGGAAGATSLCVVYPLD----FARTRLGVDIGKGPEERQFKGLGDCIMKIAKSDGIA 183
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
L++G +V +A F DT K + PK + FF+ V++ S
Sbjct 184 GLYQGFGVSVQGIIVYRASYFGAYDTVKGLLPKPKKTPFLVSFFIAQVVT-----TCSGI 238
Query 138 VVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
+ YP D R R+ G+ + R++ G DC K+ + G S ++G
Sbjct 239 LSYPFDTVRRRMMMQSGE-AKRQYKGTLDCFVKIYQHEGISSFFRG 283
Score = 34.7 bits (78), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 31/52 (59%), Gaps = 8/52 (15%)
Query 38 RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIR 89
R +M++Q+ ++ +Y G ++CF ++ + +G+ S +RG +NV+R
Sbjct 247 RRRMMMQSGEA--------KRQYKGTLDCFVKIYQHEGISSFFRGAFSNVLR 290
> hsa:293 SLC25A6, AAC3, ANT3, ANT3Y, MGC17525; solute carrier
family 25 (mitochondrial carrier; adenine nucleotide translocator),
member 6; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=298
Score = 236 bits (601), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 118/203 (58%), Positives = 150/203 (73%), Gaps = 7/203 (3%)
Query 13 IDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCK 72
I AK+F+AGG++A +SKT VAP+ERVK+L+Q Q + I +Y GI++C R+ K
Sbjct 6 ISFAKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQI--AADKQYKGIVDCIVRIPK 63
Query 73 EQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLA 131
EQGV S WRGNLANVIRYFPTQA NF FKD +K+IF +++ +F+++F N+ SGG A
Sbjct 64 EQGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHTQFWRYFAGNLASGGAA 123
Query 132 GAASLTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQG 190
GA SL VYPLDFARTRLA+DVGK G+ REF GLGDC+ K+++ G LYQGF VSVQG
Sbjct 124 GATSLCFVYPLDFARTRLAADVGKSGTEREFRGLGDCLVKITKSDGIRGLYQGFSVSVQG 183
Query 191 IIVYRGAYFGLYDAA---VPAPN 210
II+YR AYFG+YD A +P P
Sbjct 184 IIIYRAAYFGVYDTAKGMLPDPK 206
Score = 50.1 bits (118), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 39/169 (23%), Positives = 71/169 (42%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + G + G+ +C ++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKSGTEREFRGLGDCLVKITKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P + + ++ A +
Sbjct 170 IRGLYQGFSVSVQGIIIYRAAYFGVYDTAKGMLPDPKNTHIVVSWMIAQTVT-----AVA 224
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
V YP D R R+ G KG+ + G DC +K+ R G + ++G
Sbjct 225 GVVSYPFDTVRRRMMMQSGRKGADIMYTGTVDCWRKIFRDEGGKAFFKG 273
Score = 34.3 bits (77), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 44/89 (49%), Gaps = 6/89 (6%)
Query 19 FIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYS 78
IA V+A V+ + P + V+ + Q R G Y+G ++C+ ++ +++G +
Sbjct 215 MIAQTVTA-VAGVVSYPFDTVRRRMMMQSG----RKGADIMYTGTVDCWRKIFRDEGGKA 269
Query 79 LWRGNLANVIRYFPTQAFNFTFKDTFKKI 107
++G +NV+R AF D KK+
Sbjct 270 FFKGAWSNVLRGMGG-AFVLVLYDELKKV 297
> mmu:73333 Slc25a31, 1700034J06Rik, Ant4, Sfec; solute carrier
family 25 (mitochondrial carrier; adenine nucleotide translocator),
member 31; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=320
Score = 234 bits (596), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 116/201 (57%), Positives = 147/201 (73%), Gaps = 7/201 (3%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
+K+ +AGGV+A VSKT VAP+ERVK+L+Q Q S I RY G+++C R+ +EQG
Sbjct 22 SKDLLAGGVAAAVSKTAVAPIERVKLLLQVQASSKQI--SPEARYKGMLDCLVRIPREQG 79
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFKFFLTNVLSGGLAGAA 134
S WRGNLANVIRYFPTQA NF FKD +K++F N+ ++F+++FL N+ SGG AGA
Sbjct 80 FLSYWRGNLANVIRYFPTQALNFAFKDKYKELFMSGVNKEKQFWRWFLANLASGGAAGAT 139
Query 135 SLTVVYPLDFARTRLASDVGKG-SAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIV 193
SL VVYPLDFARTRL D+GKG R+F GLGDCI K+++ G + LYQGFGVSVQGIIV
Sbjct 140 SLCVVYPLDFARTRLGVDIGKGPEQRQFTGLGDCIMKIAKSDGLIGLYQGFGVSVQGIIV 199
Query 194 YRGAYFGLYDAA---VPAPNQ 211
YR +YFG YD +P P +
Sbjct 200 YRASYFGAYDTVKGLLPKPKE 220
Score = 35.4 bits (80), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 39/74 (52%), Gaps = 10/74 (13%)
Query 18 NFIAGGVSAGVSKTIVAPVERVK--MLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
+FI + S + P + V+ M++Q+ +S +Y G I+CF ++ + +G
Sbjct 226 SFIIAQIVTTCSGILSYPFDTVRRRMMMQSGES--------DRQYKGTIDCFLKIYRHEG 277
Query 76 VYSLWRGNLANVIR 89
V + +RG +N++R
Sbjct 278 VPAFFRGAFSNILR 291
> mmu:11740 Slc25a5, Ant2, MGC101927; solute carrier family 25
(mitochondrial carrier, adenine nucleotide translocator), member
5; K05863 solute carrier family 25 (mitochondrial adenine
nucleotide translocator), member 4/5/6/31
Length=298
Score = 234 bits (596), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 119/207 (57%), Positives = 149/207 (71%), Gaps = 7/207 (3%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFS 68
+ + AK+F+AGGV+A +SKT VAP+ERVK+L+Q Q + I +Y GII+C
Sbjct 2 TDAAVSFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQIT--ADKQYKGIIDCVV 59
Query 69 RVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLS 127
R+ KEQGV S WRGNLANVIRYFPTQA NF FKD +K+IF ++ +F+++F N+ S
Sbjct 60 RIPKEQGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLAS 119
Query 128 GGLAGAASLTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQGFGV 186
GG AGA SL VYPLDFARTRLA+DVGK G+ REF GLGDC+ K+ + G LYQGF V
Sbjct 120 GGAAGATSLCFVYPLDFARTRLAADVGKAGAEREFKGLGDCLVKIYKSDGIKGLYQGFNV 179
Query 187 SVQGIIVYRGAYFGLYDAA---VPAPN 210
SVQGII+YR AYFG+YD A +P P
Sbjct 180 SVQGIIIYRAAYFGIYDTAKGMLPDPK 206
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 74/169 (43%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + G + G+ +C ++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKAGAEREFKGLGDCLVKIYKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P F + + ++ +AG S
Sbjct 170 IKGLYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKNTHIFISWMIAQSVTA-VAGLTS 228
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
YP D R R+ G KG+ + G DC +K++R G + ++G
Sbjct 229 ----YPFDTVRRRMMMQSGRKGTDIMYTGTLDCWRKIARDEGSKAFFKG 273
Score = 33.9 bits (76), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 7/69 (10%)
Query 38 RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFN 97
R +M++Q+ R G Y+G ++C+ ++ +++G + ++G +NV+R AF
Sbjct 235 RRRMMMQSG------RKGTDIMYTGTLDCWRKIARDEGSKAFFKGAWSNVLRGMGG-AFV 287
Query 98 FTFKDTFKK 106
D KK
Sbjct 288 LVLYDEIKK 296
> hsa:291 SLC25A4, AAC1, ANT, ANT1, PEO2, PEO3, T1; solute carrier
family 25 (mitochondrial carrier; adenine nucleotide translocator),
member 4; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=298
Score = 233 bits (595), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 119/199 (59%), Positives = 148/199 (74%), Gaps = 7/199 (3%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K+F+AGGV+A VSKT VAP+ERVK+L+Q Q + I +Y GII+C R+ KEQG
Sbjct 10 KDFLAGGVAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKEQGF 67
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLAGAAS 135
S WRGNLANVIRYFPTQA NF FKD +K++F +++++F+++F N+ SGG AGA S
Sbjct 68 LSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLGGVDRHKQFWRYFAGNLASGGAAGATS 127
Query 136 LTVVYPLDFARTRLASDVGKGSA-REFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVY 194
L VYPLDFARTRLA+DVGKG+A REF GLGDCI K+ + G LYQGF VSVQGII+Y
Sbjct 128 LCFVYPLDFARTRLAADVGKGAAQREFHGLGDCIIKIFKSDGLRGLYQGFNVSVQGIIIY 187
Query 195 RGAYFGLYDAA---VPAPN 210
R AYFG+YD A +P P
Sbjct 188 RAAYFGVYDTAKGMLPDPK 206
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 39/169 (23%), Positives = 76/169 (44%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + + G+ +C ++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKGAAQREFHGLGDCIIKIFKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P + + F++ +++ + A
Sbjct 170 LRGLYQGFNVSVQGIIIYRAAYFGVYDTAKGMLP----DPKNVHIFVSWMIAQSVTAVAG 225
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
L V YP D R R+ G KG+ + G DC +K+++ G + ++G
Sbjct 226 L-VSYPFDTVRRRMMMQSGRKGADIMYTGTVDCWRKIAKDEGAKAFFKG 273
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 7/68 (10%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGS-----AREFLGLGDCIKKVSRRT 175
FL + L+GG+A A S T V P++ R +L V S +++ G+ DC+ ++ +
Sbjct 8 FLKDFLAGGVAAAVSKTAVAPIE--RVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQ 65
Query 176 GFLSLYQG 183
GFLS ++G
Sbjct 66 GFLSFWRG 73
Score = 36.6 bits (83), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 6/88 (6%)
Query 19 FIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYS 78
IA V+A V+ + P + V+ + Q R G Y+G ++C+ ++ K++G +
Sbjct 215 MIAQSVTA-VAGLVSYPFDTVRRRMMMQSG----RKGADIMYTGTVDCWRKIAKDEGAKA 269
Query 79 LWRGNLANVIRYFPTQAFNFTFKDTFKK 106
++G +NV+R AF D KK
Sbjct 270 FFKGAWSNVLRGMGG-AFVLVLYDEIKK 296
> mmu:11739 Slc25a4, AU019225, Ant1; solute carrier family 25
(mitochondrial carrier, adenine nucleotide translocator), member
4; K05863 solute carrier family 25 (mitochondrial adenine
nucleotide translocator), member 4/5/6/31
Length=298
Score = 233 bits (595), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 118/199 (59%), Positives = 148/199 (74%), Gaps = 7/199 (3%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K+F+AGG++A VSKT VAP+ERVK+L+Q Q + I +Y GII+C R+ KEQG
Sbjct 10 KDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKEQGF 67
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLAGAAS 135
S WRGNLANVIRYFPTQA NF FKD +K+IF +++++F+++F N+ SGG AGA S
Sbjct 68 LSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAGATS 127
Query 136 LTVVYPLDFARTRLASDVGKGSA-REFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVY 194
L VYPLDFARTRLA+DVGKGS+ REF GLGDC+ K+ + G LYQGF VSVQGII+Y
Sbjct 128 LCFVYPLDFARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGIIIY 187
Query 195 RGAYFGLYDAA---VPAPN 210
R AYFG+YD A +P P
Sbjct 188 RAAYFGVYDTAKGMLPDPK 206
Score = 46.6 bits (109), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 39/170 (22%), Positives = 78/170 (45%), Gaps = 12/170 (7%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGIINCFSRVCKEQ 74
A N +GG + S V P++ + + + G + R ++G+ +C +++ K
Sbjct 114 AGNLASGGAAGATSLCFVYPLDFARTRLAAD-----VGKGSSQREFNGLGDCLTKIFKSD 168
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G+ L++G +V +A F DT K + P + + ++ +++ + A
Sbjct 169 GLKGLYQGFSVSVQGIIIYRAAYFGVYDTAKGMLP----DPKNVHIIVSWMIAQSVTAVA 224
Query 135 SLTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
L V YP D R R+ G KG+ + G DC +K+++ G + ++G
Sbjct 225 GL-VSYPFDTVRRRMMMQSGRKGADIMYTGTLDCWRKIAKDEGANAFFKG 273
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 7/68 (10%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGS-----AREFLGLGDCIKKVSRRT 175
FL + L+GG+A A S T V P++ R +L V S +++ G+ DC+ ++ +
Sbjct 8 FLKDFLAGGIAAAVSKTAVAPIE--RVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQ 65
Query 176 GFLSLYQG 183
GFLS ++G
Sbjct 66 GFLSFWRG 73
Score = 36.2 bits (82), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 6/88 (6%)
Query 19 FIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYS 78
IA V+A V+ + P + V+ + Q R G Y+G ++C+ ++ K++G +
Sbjct 215 MIAQSVTA-VAGLVSYPFDTVRRRMMMQSG----RKGADIMYTGTLDCWRKIAKDEGANA 269
Query 79 LWRGNLANVIRYFPTQAFNFTFKDTFKK 106
++G +NV+R AF D KK
Sbjct 270 FFKGAWSNVLRGMGG-AFVLVLYDEIKK 296
> dre:192321 slc25a5, CHUNP6931, SI:bZ46J2.1, SI:bZ46J2.3, SI:bZ46J2.5,
SI:bZ46J2.6, SI:bZ46J2.7, SI:bZ46J2.9, si:bz46j2.2,
sr:nyz040, wu:fe06b05; solute carrier family 25 alpha, member
5; K05863 solute carrier family 25 (mitochondrial adenine
nucleotide translocator), member 4/5/6/31
Length=298
Score = 232 bits (592), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 117/203 (57%), Positives = 148/203 (72%), Gaps = 7/203 (3%)
Query 13 IDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCK 72
I AK+F+AGG++A +SKT VAP+ERVK+L+Q Q + I +Y GI++C R+ K
Sbjct 6 ISFAKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQIT--ADKQYKGIMDCVVRIPK 63
Query 73 EQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLA 131
EQG S WRGNLANVIRYFPTQA NF FKD +KK+F ++ +F+++F N+ SGG A
Sbjct 64 EQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKKVFLDGVDKRTQFWRYFAGNLASGGAA 123
Query 132 GAASLTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQG 190
GA SL VYPLDFARTRLA+DVGK G+ REF GLG+C+ K+S+ G LYQGF VSVQG
Sbjct 124 GATSLCFVYPLDFARTRLAADVGKAGAEREFSGLGNCLVKISKSDGIKGLYQGFNVSVQG 183
Query 191 IIVYRGAYFGLYDAA---VPAPN 210
II+YR AYFG+YD A +P P
Sbjct 184 IIIYRAAYFGIYDTAKGMLPDPK 206
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 43/169 (25%), Positives = 75/169 (44%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + G +SG+ NC ++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKAGAEREFSGLGNCLVKISKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P + + ++ +AG AS
Sbjct 170 IKGLYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKNTHIVVSWMIAQSVT-AVAGLAS 228
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
YP D R R+ G KG+ + G DC +K++R G + ++G
Sbjct 229 ----YPFDTVRRRMMMQSGRKGADIMYSGTIDCWRKIARDEGGKAFFKG 273
Score = 36.6 bits (83), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Query 38 RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFN 97
R +M++Q+ R G YSG I+C+ ++ +++G + ++G +NV+R AF
Sbjct 235 RRRMMMQSG------RKGADIMYSGTIDCWRKIARDEGGKAFFKGAWSNVLRGMGG-AFV 287
Query 98 FTFKDTFKKI 107
D KK+
Sbjct 288 LVLYDELKKV 297
> dre:327067 slc25a4, fa22e07, wu:fa22e07, zgc:77591; solute carrier
family 25 (mitochondrial carrier; adenine nucleotide
translocator), member 4; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=298
Score = 232 bits (592), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 118/208 (56%), Positives = 149/208 (71%), Gaps = 7/208 (3%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFS 68
++ I K+F+AGGV+A +SKT VAP+ERVK+L+Q Q + I +Y GII+C
Sbjct 2 SDAAISFLKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQIT--ADMQYKGIIDCVV 59
Query 69 RVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLS 127
R+ KEQG S WRGNLANVIRYFPTQA NF FKD +KKIF ++N +F+++F N+ S
Sbjct 60 RIPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKKIFLGGVDKNTQFWRYFAGNLAS 119
Query 128 GGLAGAASLTVVYPLDFARTRLASDVGKGSA-REFLGLGDCIKKVSRRTGFLSLYQGFGV 186
GG AGA SL VYPLDFARTRLA+D+GKG+A REF GLG+C+ K+ + G LY GF V
Sbjct 120 GGAAGATSLCFVYPLDFARTRLAADIGKGAAEREFTGLGNCVAKIFKSDGLRGLYLGFNV 179
Query 187 SVQGIIVYRGAYFGLYDAA---VPAPNQ 211
SVQGII+YR AYFG+YD A +P P
Sbjct 180 SVQGIIIYRAAYFGIYDTAKGMLPDPKH 207
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/170 (23%), Positives = 72/170 (42%), Gaps = 12/170 (7%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGIINCFSRVCKEQ 74
A N +GG + S V P++ + + I G R ++G+ NC +++ K
Sbjct 114 AGNLASGGAAGATSLCFVYPLDFARTRLAAD-----IGKGAAEREFTGLGNCVAKIFKSD 168
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G+ L+ G +V +A F DT K + P + + ++ A
Sbjct 169 GLRGLYLGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKHTHIVVSWMIAQTVT-----AV 223
Query 135 SLTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
+ + YP D R R+ G KG+ + G DC KK+++ G + ++G
Sbjct 224 AGIISYPFDTVRRRMMMQSGRKGADIMYKGTIDCWKKIAKDEGGKAFFKG 273
Score = 38.5 bits (88), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 43/88 (48%), Gaps = 6/88 (6%)
Query 19 FIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYS 78
IA V+A V+ I P + V+ + Q R G Y G I+C+ ++ K++G +
Sbjct 215 MIAQTVTA-VAGIISYPFDTVRRRMMMQSG----RKGADIMYKGTIDCWKKIAKDEGGKA 269
Query 79 LWRGNLANVIRYFPTQAFNFTFKDTFKK 106
++G L+NVIR AF D KK
Sbjct 270 FFKGALSNVIRG-AGGAFVLVLYDEIKK 296
> cel:K01H12.2 ant-1.3; Adenine Nucleotide Translocator family
member (ant-1.3); K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=313
Score = 232 bits (592), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 118/206 (57%), Positives = 149/206 (72%), Gaps = 6/206 (2%)
Query 4 EKRRDNNPLIDAAKNFI---AGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRY 60
EK++++ D K I +GG +A VSKT VAP+ERVK+L+Q QD+ I RY
Sbjct 11 EKKKEDKKGFDTRKFLIDLASGGTAAAVSKTAVAPIERVKLLLQVQDASLTI--AADKRY 68
Query 61 SGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFK 119
GI++ RV KEQG +LWRGNLANVIRYFPTQA NF FKDT+K IF K ++ ++F+K
Sbjct 69 KGIVDVLVRVPKEQGYAALWRGNLANVIRYFPTQALNFAFKDTYKNIFQKGLDKKKDFWK 128
Query 120 FFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLS 179
FF N+ SGG AGA SL VYPLDFARTRLA+DVGK + REF GL DC+ K+++ G +
Sbjct 129 FFAGNLASGGAAGATSLCFVYPLDFARTRLAADVGKANEREFKGLADCLVKIAKSDGPIG 188
Query 180 LYQGFGVSVQGIIVYRGAYFGLYDAA 205
LY+GF VSVQGII+YR AYFG++D A
Sbjct 189 LYRGFFVSVQGIIIYRAAYFGMFDTA 214
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 37/169 (21%), Positives = 69/169 (40%), Gaps = 12/169 (7%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ + + + + + G+ +C ++ K G
Sbjct 131 AGNLASGGAAGATSLCFVYPLDFARTRLAAD-----VGKANEREFKGLADCLVKIAKSDG 185
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFF-KFFLTNVLSGGLAGAA 134
L+RG +V +A F DT K +F + FF + + V++ G
Sbjct 186 PIGLYRGFFVSVQGIIIYRAAYFGMFDTAKMVFTADGKKLNFFAAWAIAQVVTVG----- 240
Query 135 SLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
S + YP D R R+ G+ + DC K+ + G ++++G
Sbjct 241 SGIISYPWDTVRRRMMMQSGRKDVL-YKNTLDCAVKIIKNEGMSAMFKG 288
> hsa:292 SLC25A5, 2F1, AAC2, ANT2, T2, T3; solute carrier family
25 (mitochondrial carrier; adenine nucleotide translocator),
member 5; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=298
Score = 232 bits (592), Expect = 7e-61, Method: Compositional matrix adjust.
Identities = 119/207 (57%), Positives = 148/207 (71%), Gaps = 7/207 (3%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFS 68
+ + AK+F+AGGV+A +SKT VAP+ERVK+L+Q Q + I +Y GII+C
Sbjct 2 TDAAVSFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQIT--ADKQYKGIIDCVV 59
Query 69 RVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLS 127
R+ KEQGV S WRGNLANVIRYFPTQA NF FKD +K+IF ++ +F+ +F N+ S
Sbjct 60 RIPKEQGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWLYFAGNLAS 119
Query 128 GGLAGAASLTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQGFGV 186
GG AGA SL VYPLDFARTRLA+DVGK G+ REF GLGDC+ K+ + G LYQGF V
Sbjct 120 GGAAGATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIKGLYQGFNV 179
Query 187 SVQGIIVYRGAYFGLYDAA---VPAPN 210
SVQGII+YR AYFG+YD A +P P
Sbjct 180 SVQGIIIYRAAYFGIYDTAKGMLPDPK 206
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 40/169 (23%), Positives = 73/169 (43%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + G + G+ +C ++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKAGAEREFRGLGDCLVKIYKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P + + ++ +AG S
Sbjct 170 IKGLYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKNTHIVISWMIAQTVTA-VAGLTS 228
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
YP D R R+ G KG+ + G DC +K++R G + ++G
Sbjct 229 ----YPFDTVRRRMMMQSGRKGTDIMYTGTLDCWRKIARDEGGKAFFKG 273
Score = 33.1 bits (74), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 7/69 (10%)
Query 38 RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFN 97
R +M++Q+ R G Y+G ++C+ ++ +++G + ++G +NV+R AF
Sbjct 235 RRRMMMQSG------RKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGG-AFV 287
Query 98 FTFKDTFKK 106
D KK
Sbjct 288 LVLYDEIKK 296
> dre:566370 slc25a6, si:dkey-21o13.4, wu:fj78b08; solute carrier
family 25 (mitochondrial carrier; adenine nucleotide translocator),
member 6; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=298
Score = 231 bits (590), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 117/200 (58%), Positives = 146/200 (73%), Gaps = 7/200 (3%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
AK+F+AGGV+A +SKT VAP+ERVK+L+Q Q + I +Y GI++C R+ KEQG
Sbjct 9 AKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQI--SADKQYKGIVDCIVRIPKEQG 66
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLAGAA 134
S WRGNLANVIRYFPTQA NF FKD +K+IF +++ +F+++F N+ SGG AGA
Sbjct 67 FASFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHTQFWRYFAGNLASGGAAGAT 126
Query 135 SLTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIV 193
SL VYPLDFARTRLA+DVGK GS REF GL DC+ K+ + G LYQGF VSVQGII+
Sbjct 127 SLCFVYPLDFARTRLAADVGKAGSTREFSGLADCLAKIFKSDGLRGLYQGFNVSVQGIII 186
Query 194 YRGAYFGLYDAA---VPAPN 210
YR AYFG+YD A +P P
Sbjct 187 YRAAYFGVYDTAKGMLPDPK 206
Score = 56.6 bits (135), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 75/169 (44%), Gaps = 10/169 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ +T+ + + G T +SG+ +C +++ K G
Sbjct 114 AGNLASGGAAGATSLCFVYPLD----FARTRLAADVGKAGSTREFSGLADCLAKIFKSDG 169
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ L++G +V +A F DT K + P + + ++ A +
Sbjct 170 LRGLYQGFNVSVQGIIIYRAAYFGVYDTAKGMLPDPKNTHIMVSWMIAQTVT-----AVA 224
Query 136 LTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
V YP D R R+ G KG+ + G DC +K++R G + ++G
Sbjct 225 GVVSYPFDTVRRRMMMQSGRKGADIMYTGTLDCWRKIARDEGSKAFFKG 273
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 45/88 (51%), Gaps = 6/88 (6%)
Query 19 FIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYS 78
IA V+A V+ + P + V+ + Q R G Y+G ++C+ ++ +++G +
Sbjct 215 MIAQTVTA-VAGVVSYPFDTVRRRMMMQSG----RKGADIMYTGTLDCWRKIARDEGSKA 269
Query 79 LWRGNLANVIRYFPTQAFNFTFKDTFKK 106
++G L+NV+R AF D FKK
Sbjct 270 FFKGALSNVLRGMGG-AFVLVLYDEFKK 296
Score = 38.1 bits (87), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 37/68 (54%), Gaps = 7/68 (10%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSA-----REFLGLGDCIKKVSRRT 175
F + L+GG+A A S T V P++ R +L V S +++ G+ DCI ++ +
Sbjct 8 FAKDFLAGGVAAAISKTAVAPIE--RVKLLLQVQHASKQISADKQYKGIVDCIVRIPKEQ 65
Query 176 GFLSLYQG 183
GF S ++G
Sbjct 66 GFASFWRG 73
> cel:T01B11.4 ant-1.44; Adenine Nucleotide Translocator family
member (ant-1.44); K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=313
Score = 230 bits (587), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 117/206 (56%), Positives = 149/206 (72%), Gaps = 6/206 (2%)
Query 4 EKRRDNNPLIDAAKNFI---AGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRY 60
+K++++ D K I +GG +A VSKT VAP+ERVK+L+Q QD+ I RY
Sbjct 11 DKKKEDKKGFDTRKFLIDLASGGTAAAVSKTAVAPIERVKLLLQVQDASLTI--AADKRY 68
Query 61 SGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFK 119
GI++ RV KEQG +LWRGNLANVIRYFPTQA NF FKDT+K IF K ++ ++F+K
Sbjct 69 KGIVDVLVRVPKEQGYAALWRGNLANVIRYFPTQALNFAFKDTYKNIFQKGLDKKKDFWK 128
Query 120 FFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLS 179
FF N+ SGG AGA SL VYPLDFARTRLA+DVGK + REF GL DC+ K+++ G +
Sbjct 129 FFAGNLASGGAAGATSLCFVYPLDFARTRLAADVGKANEREFKGLADCLVKIAKSDGPIG 188
Query 180 LYQGFGVSVQGIIVYRGAYFGLYDAA 205
LY+GF VSVQGII+YR AYFG++D A
Sbjct 189 LYRGFFVSVQGIIIYRAAYFGMFDTA 214
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 37/169 (21%), Positives = 69/169 (40%), Gaps = 12/169 (7%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ + + + + + G+ +C ++ K G
Sbjct 131 AGNLASGGAAGATSLCFVYPLDFARTRLAAD-----VGKANEREFKGLADCLVKIAKSDG 185
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFF-KFFLTNVLSGGLAGAA 134
L+RG +V +A F DT K +F + FF + + V++ G
Sbjct 186 PIGLYRGFFVSVQGIIIYRAAYFGMFDTAKMVFTADGKKLNFFAAWAIAQVVTVG----- 240
Query 135 SLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
S + YP D R R+ G+ + DC K+ + G ++++G
Sbjct 241 SGILSYPWDTVRRRMMMQSGRKDVL-YKNTLDCAVKIIKNEGMSAMFKG 288
> xla:432212 slc25a4, MGC79005; solute carrier family 25 (mitochondrial
carrier; adenine nucleotide translocator), member
4; K05863 solute carrier family 25 (mitochondrial adenine nucleotide
translocator), member 4/5/6/31
Length=298
Score = 230 bits (587), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 115/199 (57%), Positives = 148/199 (74%), Gaps = 7/199 (3%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K+F+AGG++A +SKT VAP+ERVK+L+Q Q + I +Y GI++C +R+ KEQG
Sbjct 10 KDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQI--SVEMQYKGIMDCVTRIPKEQGF 67
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLAGAAS 135
S WRGNLANVIRYFPTQA NF FKD +K+IF +++++F++FF+ N+ SGG AGA S
Sbjct 68 ISFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHKQFWRFFVGNLASGGAAGATS 127
Query 136 LTVVYPLDFARTRLASDVGKG-SAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVY 194
L VYPLDFARTRLA+DVGKG + REF GL +CI K+ + G LYQGF VSVQGII+Y
Sbjct 128 LCFVYPLDFARTRLAADVGKGLNEREFTGLANCIAKIYKSDGLKGLYQGFNVSVQGIIIY 187
Query 195 RGAYFGLYDAA---VPAPN 210
R AYFG+YD A +P P
Sbjct 188 RAAYFGVYDTAKGMMPDPK 206
Score = 50.1 bits (118), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 40/167 (23%), Positives = 78/167 (46%), Gaps = 10/167 (5%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
N +GG + S V P++ +T+ + + + ++G+ NC +++ K G+
Sbjct 116 NLASGGAAGATSLCFVYPLD----FARTRLAADVGKGLNEREFTGLANCIAKIYKSDGLK 171
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
L++G +V +A F DT K + P + + F++ +++ + A L
Sbjct 172 GLYQGFNVSVQGIIIYRAAYFGVYDTAKGMMP----DPKNVHIFVSWMIAQSVTAVAGL- 226
Query 138 VVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
V YP D R R+ G KG+ + G DC KK+++ G + ++G
Sbjct 227 VSYPFDTVRRRMMMQSGRKGADIMYKGTIDCWKKIAKDEGSKAFFKG 273
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 39/68 (57%), Gaps = 7/68 (10%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAR-----EFLGLGDCIKKVSRRT 175
FL + L+GG+A A S T V P++ R +L V S + ++ G+ DC+ ++ +
Sbjct 8 FLKDFLAGGIAAAISKTAVAPIE--RVKLLLQVQHASKQISVEMQYKGIMDCVTRIPKEQ 65
Query 176 GFLSLYQG 183
GF+S ++G
Sbjct 66 GFISFWRG 73
Score = 36.2 bits (82), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 42/88 (47%), Gaps = 6/88 (6%)
Query 19 FIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYS 78
IA V+A V+ + P + V+ + Q R G Y G I+C+ ++ K++G +
Sbjct 215 MIAQSVTA-VAGLVSYPFDTVRRRMMMQSG----RKGADIMYKGTIDCWKKIAKDEGSKA 269
Query 79 LWRGNLANVIRYFPTQAFNFTFKDTFKK 106
++G +NV+R AF D KK
Sbjct 270 FFKGAWSNVLRGMGG-AFVLVLYDEIKK 296
> cel:T27E9.1 ant-1.1; Adenine Nucleotide Translocator family
member (ant-1.1); K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=300
Score = 229 bits (585), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 119/205 (58%), Positives = 148/205 (72%), Gaps = 7/205 (3%)
Query 2 KEEKRRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYS 61
KE+ LID A +GG +A VSKT VAP+ERVK+L+Q QD+ AI RY
Sbjct 3 KEKSFDTKKFLIDLA----SGGTAAAVSKTAVAPIERVKLLLQVQDASKAI--AVDKRYK 56
Query 62 GIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFKF 120
GI++ RV KEQGV +LWRGNLANVIRYFPTQA NF FKDT+K IF + ++ ++F+KF
Sbjct 57 GIMDVLIRVPKEQGVAALWRGNLANVIRYFPTQAMNFAFKDTYKAIFLEGLDKKKDFWKF 116
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSL 180
F N+ SGG AGA SL VYPLDFARTRLA+D+GK + REF GL DC+ K+ + G + L
Sbjct 117 FAGNLASGGAAGATSLCFVYPLDFARTRLAADIGKANDREFKGLADCLIKIVKSDGPIGL 176
Query 181 YQGFGVSVQGIIVYRGAYFGLYDAA 205
Y+GF VSVQGII+YR AYFG++D A
Sbjct 177 YRGFFVSVQGIIIYRAAYFGMFDTA 201
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 40/169 (23%), Positives = 70/169 (41%), Gaps = 12/169 (7%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ + + I + + G+ +C ++ K G
Sbjct 118 AGNLASGGAAGATSLCFVYPLDFARTRLAAD-----IGKANDREFKGLADCLIKIVKSDG 172
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFF-LTNVLSGGLAGAA 134
L+RG +V +A F DT K +F Q FF + + V++ G
Sbjct 173 PIGLYRGFFVSVQGIIIYRAAYFGMFDTAKMVFASDGQKLNFFAAWGIAQVVTVG----- 227
Query 135 SLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
S + YP D R R+ G+ + DC KK+ + G ++++G
Sbjct 228 SGILSYPWDTVRRRMMMQSGRKDIL-YKNTLDCAKKIIQNEGMSAMFKG 275
> tgo:TGME49_100360 mitochondrial carrier domain-containing protein
; K05863 solute carrier family 25 (mitochondrial adenine
nucleotide translocator), member 4/5/6/31
Length=317
Score = 226 bits (576), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 108/192 (56%), Positives = 144/192 (75%), Gaps = 3/192 (1%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAI--RDGHTPRYSGIINCFSRVCKEQ 74
++F+ GG++ G+SKT VAPVERVK+L+Q QD+ I ++G +Y G+ +CF RV +EQ
Sbjct 24 RDFLMGGLAGGISKTFVAPVERVKLLLQLQDASTQIGHQEGQIKKYEGLKDCFVRVHREQ 83
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G+YS WRGN ANV+RYFPTQA NF K+ ++K+F +++ ++F+KFF + SGG AGA
Sbjct 84 GLYSFWRGNWANVVRYFPTQALNFACKEKYQKLFVRHDPKQDFWKFFAETLASGGAAGAT 143
Query 135 SLTVVYPLDFARTRLASDVGKGSA-REFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIV 193
SL+ VYPLDFARTRL +DVGK A R+F GL DCI+K+ + G LY+GF VSV GIIV
Sbjct 144 SLSFVYPLDFARTRLGADVGKVQAERQFTGLNDCIRKIYQEFGIPGLYRGFLVSVAGIIV 203
Query 194 YRGAYFGLYDAA 205
YR A+FGLYD A
Sbjct 204 YRAAFFGLYDTA 215
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 38/70 (54%), Gaps = 7/70 (10%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRL-----ASDVG--KGSAREFLGLGDCIKKVSR 173
FL + L GGLAG S T V P++ + L ++ +G +G +++ GL DC +V R
Sbjct 22 FLRDFLMGGLAGGISKTFVAPVERVKLLLQLQDASTQIGHQEGQIKKYEGLKDCFVRVHR 81
Query 174 RTGFLSLYQG 183
G S ++G
Sbjct 82 EQGLYSFWRG 91
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 42/84 (50%), Gaps = 9/84 (10%)
Query 6 RRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIIN 65
++ N+P++ +NF G + I P++ V+ + Q A+R Y +
Sbjct 223 KKVNHPVM---RNFAIGLGVETAAGVIAYPLDTVRRRMMMQ----ALRSDTL--YRNTWD 273
Query 66 CFSRVCKEQGVYSLWRGNLANVIR 89
C R+ +E+GV + ++G +N++R
Sbjct 274 CAGRIAREEGVAAYYKGCASNIVR 297
> cel:W02D3.6 ant-1.2; Adenine Nucleotide Translocator family
member (ant-1.2); K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=300
Score = 218 bits (556), Expect = 9e-57, Method: Compositional matrix adjust.
Identities = 110/195 (56%), Positives = 140/195 (71%), Gaps = 7/195 (3%)
Query 12 LIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVC 71
L+D A +GG +A +SKT VAP+ERVK+L+Q D + +Y GI++ +RV
Sbjct 13 LVDLA----SGGTAAAISKTAVAPIERVKLLLQVSDVSETVT--ADKKYKGIMDVLARVP 66
Query 72 KEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFKFFLTNVLSGGL 130
KEQG + WRGNLANV+RYFPTQA NF FKDT+KK+F + ++N+EF+KFF N+ SGG
Sbjct 67 KEQGYAAFWRGNLANVLRYFPTQALNFAFKDTYKKMFQEGIDKNKEFWKFFAGNLASGGA 126
Query 131 AGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQG 190
AGA SL VYPLDF RTRL +DVGKG REF GL DC K+ + G + LY+GF VSVQG
Sbjct 127 AGATSLCFVYPLDFVRTRLGADVGKGVDREFQGLTDCFVKIVKSDGPIGLYRGFFVSVQG 186
Query 191 IIVYRGAYFGLYDAA 205
II+YR AYFG++D A
Sbjct 187 IIIYRAAYFGMFDTA 201
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 42/168 (25%), Positives = 72/168 (42%), Gaps = 10/168 (5%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
A N +GG + S V P++ V+ + + G + G+ +CF ++ K G
Sbjct 118 AGNLASGGAAGATSLCFVYPLDFVRTRLGAD-----VGKGVDREFQGLTDCFVKIVKSDG 172
Query 76 VYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAAS 135
L+RG +V +A F DT K ++ Q FF + + G G+
Sbjct 173 PIGLYRGFFVSVQGIIIYRAAYFGMFDTAKTLYSTDGQKLNFFTTW--AIAQVGTVGSGY 230
Query 136 LTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
L+ YP D R R+ G+ + DC++K+ + G +LY+G
Sbjct 231 LS--YPWDTVRRRMMMQSGRKDIL-YKNTLDCVRKIVKNEGITALYKG 275
Score = 34.3 bits (77), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 118 FKFFLTNVLSGGLAGAASLTVVYPLDFARTRL-ASDVGK--GSAREFLGLGDCIKKVSRR 174
++ FL ++ SGG A A S T V P++ + L SDV + + +++ G+ D + +V +
Sbjct 9 YRKFLVDLASGGTAAAISKTAVAPIERVKLLLQVSDVSETVTADKKYKGIMDVLARVPKE 68
Query 175 TGFLSLYQG 183
G+ + ++G
Sbjct 69 QGYAAFWRG 77
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 10/30 (33%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 60 YSGIINCFSRVCKEQGVYSLWRGNLANVIR 89
Y ++C ++ K +G+ +L++G L+NV R
Sbjct 253 YKNTLDCVRKIVKNEGITALYKGGLSNVFR 282
> dre:100038774 si:dkey-251i10.1; K05863 solute carrier family
25 (mitochondrial adenine nucleotide translocator), member
4/5/6/31
Length=299
Score = 208 bits (530), Expect = 8e-54, Method: Compositional matrix adjust.
Identities = 115/208 (55%), Positives = 150/208 (72%), Gaps = 8/208 (3%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFS 68
++ ++ AK+F+AGGV+A VS+T VAP+ERVK+L+Q Q + I +Y GI++C
Sbjct 2 SDSVVSFAKDFLAGGVAAAVSETAVAPIERVKLLLQVQHASKQITT--DKQYKGIVDCVV 59
Query 69 RVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFKFFLTNVLS 127
R+ +EQG S WRGNLANVIRYF TQA NF FKD ++KIF +Q +F+++F N+ +
Sbjct 60 RIPREQGFLSFWRGNLANVIRYFLTQALNFAFKDKYRKIFLDGVDQRTQFWRYFAGNLAA 119
Query 128 GGLAGAASLTVVYPLDFARTRLASDVGK-GSA-REFLGLGDCIKKVSRRTGFLSLYQGFG 185
GG AGA SL +VYPLDFARTRLA+DVGK G+A REF GL +C+ K+ R G LYQGF
Sbjct 120 GGAAGATSLCLVYPLDFARTRLAADVGKTGTAGREFKGLANCLVKIYRSDGVRGLYQGFN 179
Query 186 VSVQGIIVYRGAYFGLYDAA---VPAPN 210
VSVQGII+YR AYFG+YD A +P P
Sbjct 180 VSVQGIIIYRAAYFGIYDTAKGMLPDPE 207
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 56/125 (44%), Gaps = 6/125 (4%)
Query 60 YSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFK 119
+ G+ NC ++ + GV L++G +V +A F DT K + P
Sbjct 155 FKGLANCLVKIYRSDGVRGLYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPENTHIAVS 214
Query 120 FFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFL 178
+ + + ++ +AG S YP D R + G KG+ + G DC +K++R G
Sbjct 215 WMIAHTVTI-VAGFIS----YPFDTVRRSMMMQSGLKGADVMYSGSIDCWRKIARNEGPK 269
Query 179 SLYQG 183
+ ++G
Sbjct 270 AFFKG 274
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 26/101 (25%), Positives = 44/101 (43%), Gaps = 10/101 (9%)
Query 12 LIDAAKNFIAGGVSAGVSKTIVA-----PVERVKMLIQTQDSIPAIRDGHTPRYSGIINC 66
L D IA + TIVA P + V+ + Q + G YSG I+C
Sbjct 203 LPDPENTHIAVSWMIAHTVTIVAGFISYPFDTVRRSMMMQSGLK----GADVMYSGSIDC 258
Query 67 FSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKI 107
+ ++ + +G + ++G +NV+R + D FKK+
Sbjct 259 WRKIARNEGPKAFFKGAWSNVLRSLGG-SIVLVLYDEFKKV 298
> cel:C47E12.2 hypothetical protein; K05863 solute carrier family
25 (mitochondrial adenine nucleotide translocator), member
4/5/6/31
Length=306
Score = 205 bits (522), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 106/205 (51%), Positives = 136/205 (66%), Gaps = 12/205 (5%)
Query 4 EKRRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDS---IPAIRDGHTPRY 60
+KR D A + + GGVSA VSKT+VAP+ERVK+L+Q Q S IPA + RY
Sbjct 13 QKRSDAQKF---AIDLLIGGVSASVSKTVVAPIERVKILLQVQYSHKDIPADK-----RY 64
Query 61 SGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEE-FFK 119
+GII+ F RV KEQG S WRGN+ NVIRYFPTQAFNF F D +K I K + E
Sbjct 65 NGIIDAFVRVPKEQGFVSFWRGNMTNVIRYFPTQAFNFAFNDLYKSILLKNMKRENNVLS 124
Query 120 FFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLS 179
+ + ++SGGLAG +SL +VYPLDF RTRL++D+ + RE+ GL DC K + GF +
Sbjct 125 YSVRTLVSGGLAGCSSLCIVYPLDFIRTRLSADINHHTKREYKGLVDCTMKTVKNEGFSA 184
Query 180 LYQGFGVSVQGIIVYRGAYFGLYDA 204
LY+GF +S+Q +YR YFGLYDA
Sbjct 185 LYRGFAISLQTYFIYRSVYFGLYDA 209
Score = 56.2 bits (134), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 43/179 (24%), Positives = 86/179 (48%), Gaps = 15/179 (8%)
Query 6 RRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGII 64
+R+NN L + + ++GG++ S IV P++ I+T+ + A + HT R Y G++
Sbjct 117 KRENNVLSYSVRTLVSGGLAGCSSLCIVYPLD----FIRTR--LSADINHHTKREYKGLV 170
Query 65 NCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTN 124
+C + K +G +L+RG ++ YF ++ F D + N +++ F+ +
Sbjct 171 DCTMKTVKNEGFSALYRGFAISLQTYFIYRSVYFGLYDAIRNTI---NTDKKKLPFYASF 227
Query 125 VLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
++ G+ +S + YP D R R+ +++ F +K+ G LY+G
Sbjct 228 AIAQGVTVLSSY-LTYPWDTVRRRMMVKGQLSTSKAF----SAARKIVHEEGVRGLYKG 281
> sce:YBR085W AAC3, ANC3; Aac3p; K05863 solute carrier family
25 (mitochondrial adenine nucleotide translocator), member 4/5/6/31
Length=307
Score = 201 bits (512), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 108/209 (51%), Positives = 140/209 (66%), Gaps = 11/209 (5%)
Query 3 EEKRRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHT-PRYS 61
+ K+++ N I NF+ GGVSA ++KT +P+ERVK+LIQ QD + I+ G +YS
Sbjct 4 DAKQQETNFAI----NFLMGGVSAAIAKTAASPIERVKILIQNQDEM--IKQGTLDKKYS 57
Query 62 GIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFF 121
GI++CF R K++G+ S WRGN ANVIRYFPTQA NF FKD K +F + + E + K+F
Sbjct 58 GIVDCFKRTAKQEGLISFWRGNTANVIRYFPTQALNFAFKDKIKLMFG-FKKEEGYGKWF 116
Query 122 LTNVLSGGLAGAASLTVVYPLDFARTRLASDV---GKGSAREFLGLGDCIKKVSRRTGFL 178
N+ SGG AGA SL VY LDFARTRLA+D KG AR+F GL D KK + G
Sbjct 117 AGNLASGGAAGALSLLFVYSLDFARTRLAADAKSSKKGGARQFNGLTDVYKKTLKSDGIA 176
Query 179 SLYQGFGVSVQGIIVYRGAYFGLYDAAVP 207
LY+GF SV GI+VYRG YFG++D+ P
Sbjct 177 GLYRGFMPSVVGIVVYRGLYFGMFDSLKP 205
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 42/175 (24%), Positives = 80/175 (45%), Gaps = 12/175 (6%)
Query 16 AKNFIAGGVSAGVSKTIVAPVE--RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKE 73
A N +GG + +S V ++ R ++ + S + G +++G+ + + + K
Sbjct 117 AGNLASGGAAGALSLLFVYSLDFARTRLAADAKSS----KKGGARQFNGLTDVYKKTLKS 172
Query 74 QGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGA 133
G+ L+RG + +V+ + F D+ K + + + F FL G +
Sbjct 173 DGIAGLYRGFMPSVVGIVVYRGLYFGMFDSLKPLVLTGSLDGSFLASFLL----GWVVTT 228
Query 134 ASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSV 188
+ T YPLD R R+ + G A ++ G DC+KK+ G SL++G G ++
Sbjct 229 GASTCSYPLDTVRRRMM--MTSGQAVKYNGAIDCLKKIVASEGVGSLFKGCGANI 281
Score = 36.2 bits (82), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 7/72 (9%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+F+ G V + T P++ V+ + + G +Y+G I+C ++ +GV
Sbjct 219 SFLLGWVVTTGASTCSYPLDTVRRRMM-------MTSGQAVKYNGAIDCLKKIVASEGVG 271
Query 78 SLWRGNLANVIR 89
SL++G AN++R
Sbjct 272 SLFKGCGANILR 283
> sce:YBL030C PET9, AAC2, ANC2; Major ADP/ATP carrier of the mitochondrial
inner membrane, exchanges cytosolic ADP for mitochondrially
synthesized ATP; also imports heme and ATP; phosphorylated;
required for viability in many lab strains that
carry a sal1 mutation; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=318
Score = 199 bits (506), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 108/204 (52%), Positives = 137/204 (67%), Gaps = 11/204 (5%)
Query 8 DNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGIINC 66
++N LID F+ GGVSA V+KT +P+ERVK+LIQ QD + ++ G R Y+GI++C
Sbjct 20 ESNFLID----FLMGGVSAAVAKTAASPIERVKLLIQNQDEM--LKQGTLDRKYAGILDC 73
Query 67 FSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVL 126
F R ++GV S WRGN ANVIRYFPTQA NF FKD K +F + + E + K+F N+
Sbjct 74 FKRTATQEGVISFWRGNTANVIRYFPTQALNFAFKDKIKAMFG-FKKEEGYAKWFAGNLA 132
Query 127 SGGLAGAASLTVVYPLDFARTRLASD---VGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
SGG AGA SL VY LD+ARTRLA+D KG AR+F GL D KK + G LY+G
Sbjct 133 SGGAAGALSLLFVYSLDYARTRLAADSKSSKKGGARQFNGLIDVYKKTLKSDGVAGLYRG 192
Query 184 FGVSVQGIIVYRGAYFGLYDAAVP 207
F SV GI+VYRG YFG+YD+ P
Sbjct 193 FLPSVVGIVVYRGLYFGMYDSLKP 216
Score = 59.3 bits (142), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 12/175 (6%)
Query 16 AKNFIAGGVSAGVSKTIVAPVE--RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKE 73
A N +GG + +S V ++ R ++ ++ S + G +++G+I+ + + K
Sbjct 128 AGNLASGGAAGALSLLFVYSLDYARTRLAADSKSS----KKGGARQFNGLIDVYKKTLKS 183
Query 74 QGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGA 133
GV L+RG L +V+ + F D+ K + + F FL G +
Sbjct 184 DGVAGLYRGFLPSVVGIVVYRGLYFGMYDSLKPLLLTGSLEGSFLASFLL----GWVVTT 239
Query 134 ASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSV 188
+ T YPLD R R+ + G A ++ G DC++K+ G SL++G G ++
Sbjct 240 GASTCSYPLDTVRRRMM--MTSGQAVKYDGAFDCLRKIVAAEGVGSLFKGCGANI 292
Score = 33.9 bits (76), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 35/72 (48%), Gaps = 7/72 (9%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+F+ G V + T P++ V+ + + G +Y G +C ++ +GV
Sbjct 230 SFLLGWVVTTGASTCSYPLDTVRRRMM-------MTSGQAVKYDGAFDCLRKIVAAEGVG 282
Query 78 SLWRGNLANVIR 89
SL++G AN++R
Sbjct 283 SLFKGCGANILR 294
> sce:YMR056C AAC1; Aac1p; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=309
Score = 189 bits (481), Expect = 5e-48, Method: Compositional matrix adjust.
Identities = 98/194 (50%), Positives = 126/194 (64%), Gaps = 6/194 (3%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHT-PRYSGIINCFSRVCKEQGV 76
+F+ GGVSA ++KT AP+ERVK+L+Q Q+ + ++ G RY GI++CF R +G+
Sbjct 16 DFLMGGVSAAIAKTGAAPIERVKLLMQNQEEM--LKQGSLDTRYKGILDCFKRTATHEGI 73
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
S WRGN ANV+RYFPTQA NF FKD K + + + + K+F N+ SGG AG SL
Sbjct 74 VSFWRGNTANVLRYFPTQALNFAFKDKIKSLLSYDRERDGYAKWFAGNLFSGGAAGGLSL 133
Query 137 TVVYPLDFARTRLASDV---GKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIV 193
VY LD+ARTRLA+D S R+F GL D KK + G L LY+GF SV GIIV
Sbjct 134 LFVYSLDYARTRLAADARGSKSTSQRQFNGLLDVYKKTLKTDGLLGLYRGFVPSVLGIIV 193
Query 194 YRGAYFGLYDAAVP 207
YRG YFGLYD+ P
Sbjct 194 YRGLYFGLYDSFKP 207
Score = 56.2 bits (134), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 41/175 (23%), Positives = 81/175 (46%), Gaps = 12/175 (6%)
Query 16 AKNFIAGGVSAGVSKTIVAPVE--RVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKE 73
A N +GG + G+S V ++ R ++ + S + +++G+++ + + K
Sbjct 119 AGNLFSGGAAGGLSLLFVYSLDYARTRLAADARGS----KSTSQRQFNGLLDVYKKTLKT 174
Query 74 QGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGA 133
G+ L+RG + +V+ + F D+FK + F FL G +
Sbjct 175 DGLLGLYRGFVPSVLGIIVYRGLYFGLYDSFKPVLLTGALEGSFVASFLL----GWVITM 230
Query 134 ASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSV 188
+ T YPLD R R+ + G ++ G DC++K+ ++ G SL++G G ++
Sbjct 231 GASTASYPLDTVRRRMM--MTSGQTIKYDGALDCLRKIVQKEGAYSLFKGCGANI 283
Score = 38.5 bits (88), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 38/72 (52%), Gaps = 7/72 (9%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+F+ G V + T P++ V+ + + G T +Y G ++C ++ +++G Y
Sbjct 221 SFLLGWVITMGASTASYPLDTVRRRMM-------MTSGQTIKYDGALDCLRKIVQKEGAY 273
Query 78 SLWRGNLANVIR 89
SL++G AN+ R
Sbjct 274 SLFKGCGANIFR 285
> ath:AT4G28390 AAC3; AAC3 (ADP/ATP CARRIER 3); ATP:ADP antiporter/
binding; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=379
Score = 185 bits (469), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 104/194 (53%), Positives = 134/194 (69%), Gaps = 6/194 (3%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGIINCFSRVCKEQGV 76
+F+ GGVSA VSKT AP+ERVK+LIQ QD + I+ G Y GI +CF+R K++G+
Sbjct 82 DFLMGGVSAAVSKTAAAPIERVKLLIQNQDEM--IKAGRLSEPYKGISDCFARTVKDEGM 139
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
+LWRGN ANVIRYFPTQA NF FKD FK++F + + ++K+F N+ SGG AGA+SL
Sbjct 140 LALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKEKDGYWKWFAGNLASGGAAGASSL 199
Query 137 TVVYPLDFARTRLASD---VGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIV 193
VY LD+ARTRLA+D KG R+F G+ D KK G + LY+GF +S GI+V
Sbjct 200 LFVYSLDYARTRLANDAKAAKKGGQRQFNGMVDVYKKTIASDGIVGLYRGFNISCVGIVV 259
Query 194 YRGAYFGLYDAAVP 207
YRG YFGLYD+ P
Sbjct 260 YRGLYFGLYDSLKP 273
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 10/66 (15%)
Query 24 VSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWRGN 83
+ AG++ + V R M+ G +Y + FS++ K +G SL++G
Sbjct 296 IGAGLASYPIDTVRRRMMMT----------SGEAVKYKSSLQAFSQIVKNEGAKSLFKGA 345
Query 84 LANVIR 89
AN++R
Sbjct 346 GANILR 351
Score = 30.8 bits (68), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 34/67 (50%), Gaps = 4/67 (5%)
Query 121 FLTNVLSGGLAGAASLTVVYPLDFARTRLASD---VGKGSARE-FLGLGDCIKKVSRRTG 176
FL + L GG++ A S T P++ + + + + G E + G+ DC + + G
Sbjct 79 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGISDCFARTVKDEG 138
Query 177 FLSLYQG 183
L+L++G
Sbjct 139 MLALWRG 145
> ath:AT5G13490 AAC2; AAC2 (ADP/ATP carrier 2); ATP:ADP antiporter/
binding; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=385
Score = 184 bits (467), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 106/197 (53%), Positives = 133/197 (67%), Gaps = 7/197 (3%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGH-TPRYSGIINCFSRVCKEQ 74
A +F+ GGVSA VSKT AP+ERVK+LIQ QD + ++ G T Y GI +CF R +++
Sbjct 85 AIDFMMGGVSAAVSKTAAAPIERVKLLIQNQDEM--LKAGRLTEPYKGIRDCFGRTIRDE 142
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G+ SLWRGN ANVIRYFPTQA NF FKD FK++F + ++K+F N+ SGG AGA+
Sbjct 143 GIGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAGAS 202
Query 135 SLTVVYPLDFARTRLASDVGK----GSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQG 190
SL VY LD+ARTRLA+D G R+F GL D KK + G LY+GF +S G
Sbjct 203 SLLFVYSLDYARTRLANDSKSAKKGGGERQFNGLVDVYKKTLKSDGIAGLYRGFNISCAG 262
Query 191 IIVYRGAYFGLYDAAVP 207
IIVYRG YFGLYD+ P
Sbjct 263 IIVYRGLYFGLYDSVKP 279
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 52 IRDGHTPRYSGIINCFSRVCKEQGVYSLWRGNLANVIR 89
+ G +Y + FS++ K++G SL++G AN++R
Sbjct 320 MTSGEAVKYKSSFDAFSQIVKKEGAKSLFKGAGANILR 357
> ath:AT3G08580 AAC1; AAC1 (ADP/ATP CARRIER 1); ATP:ADP antiporter/
binding; K05863 solute carrier family 25 (mitochondrial
adenine nucleotide translocator), member 4/5/6/31
Length=381
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 106/197 (53%), Positives = 131/197 (66%), Gaps = 7/197 (3%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGIINCFSRVCKEQ 74
A +F+ GGVSA VSKT AP+ERVK+LIQ QD + I+ G Y GI +CF R K++
Sbjct 81 ALDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEM--IKAGRLSEPYKGIGDCFGRTIKDE 138
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G SLWRGN ANVIRYFPTQA NF FKD FK++F + ++K+F N+ SGG AGA+
Sbjct 139 GFGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAAGAS 198
Query 135 SLTVVYPLDFARTRLASD----VGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQG 190
SL VY LD+ARTRLA+D G R+F GL D +K + G LY+GF +S G
Sbjct 199 SLLFVYSLDYARTRLANDAKAAKKGGGGRQFDGLVDVYRKTLKTDGIAGLYRGFNISCVG 258
Query 191 IIVYRGAYFGLYDAAVP 207
IIVYRG YFGLYD+ P
Sbjct 259 IIVYRGLYFGLYDSVKP 275
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 63/131 (48%), Gaps = 8/131 (6%)
Query 59 RYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFF 118
++ G+++ + + K G+ L+RG + + + F D+ K + + + FF
Sbjct 228 QFDGLVDVYRKTLKTDGIAGLYRGFNISCVGIIVYRGLYFGLYDSVKPVLLTGDLQDSFF 287
Query 119 -KFFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGF 177
F L V++ G AG AS YP+D R R+ + G A ++ D K++ + G
Sbjct 288 ASFALGWVITNG-AGLAS----YPIDTVRRRMM--MTSGEAVKYKSSLDAFKQILKNEGA 340
Query 178 LSLYQGFGVSV 188
SL++G G ++
Sbjct 341 KSLFKGAGANI 351
Score = 32.7 bits (73), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 37/73 (50%), Gaps = 4/73 (5%)
Query 115 EEFFKFFLTNVLSGGLAGAASLTVVYPLDFARTRLASD---VGKGSARE-FLGLGDCIKK 170
E+ F F + L GG++ A S T P++ + + + + G E + G+GDC +
Sbjct 74 EKGFTNFALDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGIGDCFGR 133
Query 171 VSRRTGFLSLYQG 183
+ GF SL++G
Sbjct 134 TIKDEGFGSLWRG 146
> cel:F25B4.7 hypothetical protein; K05863 solute carrier family
25 (mitochondrial adenine nucleotide translocator), member
4/5/6/31
Length=306
Score = 173 bits (438), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 88/206 (42%), Positives = 133/206 (64%), Gaps = 6/206 (2%)
Query 7 RDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINC 66
RD+ +I +K+F+AG +A +SKT++APVERVK+++Q Q+S + RY GI++C
Sbjct 12 RDD--VIKFSKDFMAGATAAAISKTVIAPVERVKLILQLQNSQTTL--ALENRYKGIVDC 67
Query 67 FSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKI-FPKYNQNEEFFKFFLTNV 125
F RV +EQG S WRGN N++R ++ +FK+ F+K + + ++ + N+
Sbjct 68 FIRVPREQGFLSFWRGNWVNILRSCSQESLGLSFKEFFRKYSLEGVDPKTQHSRWLVGNL 127
Query 126 LSGGLAGAASLTVVYPLDFARTRLASDVGK-GSAREFLGLGDCIKKVSRRTGFLSLYQGF 184
++GG +G A+L +YPLDF RTRLA D+GK S REF G+ DC KK+ + G LY+G
Sbjct 128 VAGGGSGCATLATIYPLDFIRTRLAIDLGKRKSDREFTGMFDCAKKIIKSDGVPGLYKGL 187
Query 185 GVSVQGIIVYRGAYFGLYDAAVPAPN 210
S+Q +I+YRGAY+GL+D P N
Sbjct 188 IPSLQYMIIYRGAYYGLFDTTAPYMN 213
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/170 (22%), Positives = 70/170 (41%), Gaps = 18/170 (10%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
N +AGG S + + P++ I+T+ +I + ++G+ +C ++ K GV
Sbjct 126 NLVAGGGSGCATLATIYPLD----FIRTRLAIDLGKRKSDREFTGMFDCAKKIIKSDGVP 181
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIF----PKYNQNEEFFKFFLTNVLSGGLAGA 133
L++G P+ + ++ + +F P N + + F L G +
Sbjct 182 GLYKG-------LIPSLQYMIIYRGAYYGLFDTTAPYMNSDGKMT--FTEAFLVGQVVTL 232
Query 134 ASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
+ YPLD R RL GK + F CIK + + G + + G
Sbjct 233 IAAMTSYPLDTVRRRLMMGAGKKTL-PFNNTISCIKYIYTKEGPKAFFHG 281
> ath:AT5G17400 ER-ANT1 (ENDOPLASMIC RETICULUM-DENINE NUCLEOTIDE
TRANSPORTER 1); ATP:ADP antiporter/ binding
Length=306
Score = 172 bits (435), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 93/196 (47%), Positives = 127/196 (64%), Gaps = 6/196 (3%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPR-YSGIINCFSRVCKEQ 74
+ +F+ GG +A V+K+ AP+ERVK+L+Q Q + I+ GH R Y+G+ NCF+R+ +E+
Sbjct 11 SADFVMGGAAAIVAKSAAAPIERVKLLLQNQGEM--IKTGHLIRPYTGLGNCFTRIYREE 68
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
GV S WRGN ANVIRYFPTQA NF FK FK + + + + K+F NV SG AGA
Sbjct 69 GVLSFWRGNQANVIRYFPTQASNFAFKGYFKNLLGCSKEKDGYLKWFAGNVASGSAAGAT 128
Query 135 SLTVVYPLDFARTRLASDVGKGSA---REFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGI 191
+ +Y LD+ARTRL +D + S R+F G+ D +K G LY+GFGVS+ GI
Sbjct 129 TSLFLYHLDYARTRLGTDAKECSVNGKRQFKGMIDVYRKTLSSDGIKGLYRGFGVSIVGI 188
Query 192 IVYRGAYFGLYDAAVP 207
+YRG YFG+YD P
Sbjct 189 TLYRGMYFGMYDTIKP 204
Score = 34.3 bits (77), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 27/125 (21%), Positives = 55/125 (44%), Gaps = 6/125 (4%)
Query 59 RYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFF 118
++ G+I+ + + G+ L+RG +++ + F DT K I + F
Sbjct 157 QFKGMIDVYRKTLSSDGIKGLYRGFGVSIVGITLYRGMYFGMYDTIKPIVLVGSLEGNFL 216
Query 119 KFFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFL 178
FL L + +A + + YP D R R+ + G ++ ++++ + GF
Sbjct 217 ASFL---LGWSITTSAGV-IAYPFDTLRRRMM--LTSGQPVKYRNTIHALREILKSEGFY 270
Query 179 SLYQG 183
+LY+G
Sbjct 271 ALYRG 275
> ath:AT5G56450 mitochondrial substrate carrier family protein;
K05863 solute carrier family 25 (mitochondrial adenine nucleotide
translocator), member 4/5/6/31
Length=330
Score = 168 bits (426), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 87/209 (41%), Positives = 128/209 (61%), Gaps = 11/209 (5%)
Query 6 RRDNNPLI------DAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAI--RDGHT 57
RR+ +PL K+ +AG V GV TIVAP+ER K+L+QTQ+S AI +GH
Sbjct 15 RRNQSPLSLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNIAIVGDEGHA 74
Query 58 --PRYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNE 115
R+ G+ + R +E+GV SLWRGN ++V+RY+P+ A NF+ KD ++ I + E
Sbjct 75 GKRRFKGMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRSILRNSSSQE 134
Query 116 -EFFKFFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRR 174
F L N ++G AG +L VVYPLD A TRLA+D+GK AR+F G+ + + ++
Sbjct 135 NHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPEARQFRGIHHFLSTIHKK 194
Query 175 TGFLSLYQGFGVSVQGIIVYRGAYFGLYD 203
G +Y+G S+ G+I++RG YFG +D
Sbjct 195 DGVRGIYRGLPASLHGVIIHRGLYFGGFD 223
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 47/185 (25%), Positives = 77/185 (41%), Gaps = 15/185 (8%)
Query 2 KEEKRRDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYS 61
+ ++N+ A NF+AG + + +V P++ + P R ++
Sbjct 128 RNSSSQENHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPEAR-----QFR 182
Query 62 GIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFF 121
GI + S + K+ GV ++RG A++ + F DT K+IF + + E
Sbjct 183 GIHHFLSTIHKKDGVRGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPE------ 236
Query 122 LTNVLSGGLAGAASLT---VVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFL 178
L GLA A + + YPLD R R+ G + DC KK+ R G
Sbjct 237 LALWKRWGLAQAVTTSAGLASYPLDTVRRRIMMQSGMEHPM-YRSTLDCWKKIYRSEGLA 295
Query 179 SLYQG 183
S Y+G
Sbjct 296 SFYRG 300
Score = 35.0 bits (79), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 50/110 (45%), Gaps = 13/110 (11%)
Query 2 KEEKRRDNNPLIDAAKNF-IAGGV--SAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTP 58
KE D P + K + +A V SAG++ P++ V+ I Q + P
Sbjct 226 KEIFSEDTKPELALWKRWGLAQAVTTSAGLASY---PLDTVRRRIMMQSGM------EHP 276
Query 59 RYSGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF 108
Y ++C+ ++ + +G+ S +RG L+N+ R + A F D K+
Sbjct 277 MYRSTLDCWKKIYRSEGLASFYRGALSNMFRSTGSAAI-LVFYDEVKRFL 325
> cel:R07E3.4 hypothetical protein
Length=298
Score = 155 bits (393), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 77/194 (39%), Positives = 117/194 (60%), Gaps = 5/194 (2%)
Query 12 LIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVC 71
+++ +K +AG +A +SKT AP +RVK+++Q Q + Y+GI +C S++
Sbjct 10 VLETSKKCLAGSAAAAISKTTTAPFDRVKLVLQLQRQ----SEFAMAEYNGIRDCISKIR 65
Query 72 KEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPK-YNQNEEFFKFFLTNVLSGGL 130
EQG +LWRGN A V R P NF F+D ++ K ++NE F KF +SGGL
Sbjct 66 LEQGAMALWRGNGAGVARCLPNHTLNFAFRDIYRNTLLKNVDRNESFGKFLAGTFVSGGL 125
Query 131 AGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQG 190
GA +L ++YP DFARTRLA DV K +R++ G+ DC+KK+ G S Y+G ++Q
Sbjct 126 GGATTLFMLYPFDFARTRLALDVKKDGSRKYKGMVDCLKKIKASEGVASWYKGLSSALQF 185
Query 191 IIVYRGAYFGLYDA 204
+I R +FG++D+
Sbjct 186 VIASRAIFFGIFDS 199
> xla:398474 slc25a24-a, MGC132369, apc1, scamc-1, scamc1-A, slc25a24;
solute carrier family 25 (mitochondrial carrier; phosphate
carrier), member 24; K14684 solute carrier family 25
(mitochondrial phosphate transporter), member 23/24/25/41
Length=473
Score = 112 bits (279), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 66/191 (34%), Positives = 103/191 (53%), Gaps = 25/191 (13%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K+ +AGG++ VS+T AP++R+K+++Q T S II ++ KE GV
Sbjct 196 KHLLAGGMAGAVSRTGTAPLDRLKVMMQVHG---------TKGNSNIITGLKQMVKEGGV 246
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFP----KYNQNEEFFKFFLTNVLSGGLAG 132
SLWRGN NVI+ P A F + +KK+F K E F ++G LAG
Sbjct 247 RSLWRGNGVNVIKIAPETAMKFWAYEQYKKLFTSESGKLGTAERF--------IAGSLAG 298
Query 133 AASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGII 192
A + T +YP++ +TRLA G ++ G+ DC KK+ ++ G L+ Y+G+ ++ GII
Sbjct 299 ATAQTSIYPMEVLKTRLAV----GKTGQYSGMFDCAKKIMQKEGILAFYKGYIPNILGII 354
Query 193 VYRGAYFGLYD 203
Y G +Y+
Sbjct 355 PYAGIDLAIYE 365
Score = 63.5 bits (153), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 38/172 (22%), Positives = 78/172 (45%), Gaps = 10/172 (5%)
Query 13 IDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCK 72
+ A+ FIAG ++ ++T + P+E +K + G T +YSG+ +C ++ +
Sbjct 285 LGTAERFIAGSLAGATAQTSIYPMEVLKTRLAV---------GKTGQYSGMFDCAKKIMQ 335
Query 73 EQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAG 132
++G+ + ++G + N++ P + +T K + + + L + G ++
Sbjct 336 KEGILAFYKGYIPNILGIIPYAGIDLAIYETLKNYWLQNYAKDSANPGVLVLLGCGTVSS 395
Query 133 AASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGF 184
YPL RTR+ + A + L +G +K+ + GF LY G
Sbjct 396 TCGQLASYPLALIRTRMQAQASIEGAPQ-LNMGGLFRKIVAKEGFFGLYTGI 446
> hsa:29957 SLC25A24, APC1, DKFZp586G0123, SCAMC-1; solute carrier
family 25 (mitochondrial carrier; phosphate carrier), member
24; K14684 solute carrier family 25 (mitochondrial phosphate
transporter), member 23/24/25/41
Length=477
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 61/187 (32%), Positives = 100/187 (53%), Gaps = 16/187 (8%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ +AGG++ VS+T AP++R+K+++Q G I F ++ KE G+
Sbjct 196 RQLLAGGIAGAVSRTSTAPLDRLKIMMQVH--------GSKSDKMNIFGGFRQMVKEGGI 247
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
SLWRGN NVI+ P A F + +KK+ + Q F+ F +SG +AGA +
Sbjct 248 RSLWRGNGTNVIKIAPETAVKFWAYEQYKKLLTEEGQKIGTFERF----ISGSMAGATAQ 303
Query 137 TVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRG 196
T +YP++ +TRLA G ++ G+ DC KK+ + G + Y+G+ ++ GII Y G
Sbjct 304 TFIYPMEVMKTRLAV----GKTGQYSGIYDCAKKILKHEGLGAFYKGYVPNLLGIIPYAG 359
Query 197 AYFGLYD 203
+Y+
Sbjct 360 IDLAVYE 366
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 38/188 (20%), Positives = 83/188 (44%), Gaps = 12/188 (6%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ FI+G ++ ++T + P+E +K + G T +YSGI +C ++ K +G+
Sbjct 290 ERFISGSMAGATAQTFIYPMEVMKTRLAV---------GKTGQYSGIYDCAKKILKHEGL 340
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
+ ++G + N++ P + + K + + + + G L+
Sbjct 341 GAFYKGYVPNLLGIIPYAGIDLAVYELLKSYWLDNFAKDSVNPGVMVLLGCGALSSTCGQ 400
Query 137 TVVYPLDFARTRL-ASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYR 195
YPL RTR+ A + +GS + L + +++ + G LY+G + ++
Sbjct 401 LASYPLALVRTRMQAQAMLEGSPQ--LNMVGLFRRIISKEGIPGLYRGITPNFMKVLPAV 458
Query 196 GAYFGLYD 203
G + +Y+
Sbjct 459 GISYVVYE 466
Score = 34.7 bits (78), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 29/121 (23%), Positives = 58/121 (47%), Gaps = 5/121 (4%)
Query 83 NLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLTVVYPL 142
++ +IR++ + D+ I ++ ++E+ + +L+GG+AGA S T PL
Sbjct 158 DIEEIIRFWK-HSTGIDIGDSLT-IPDEFTEDEKKSGQWWRQLLAGGIAGAVSRTSTAPL 215
Query 143 DFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRGAYFGLY 202
D R ++ V GS + + + +++ + G SL++G G +V I F Y
Sbjct 216 D--RLKIMMQV-HGSKSDKMNIFGGFRQMVKEGGIRSLWRGNGTNVIKIAPETAVKFWAY 272
Query 203 D 203
+
Sbjct 273 E 273
> dre:447867 slc25a24; zgc:92470; K14684 solute carrier family
25 (mitochondrial phosphate transporter), member 23/24/25/41
Length=477
Score = 108 bits (269), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 66/191 (34%), Positives = 102/191 (53%), Gaps = 22/191 (11%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K AGGV+ VS+T AP++R+K+ +Q S T + S ++N F ++ KE GV
Sbjct 197 KQLAAGGVAGAVSRTGTAPLDRMKVFMQVHSS-------KTNKIS-LVNGFKQMIKEGGV 248
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYN---QNEEFFKFFLTNVLSGGLAGA 133
SLWRGN NVI+ P A F + +KK+ K Q+ E F ++G LAGA
Sbjct 249 ASLWRGNGVNVIKIAPETAIKFMAYEQYKKLLSKDGGKVQSHERF-------MAGSLAGA 301
Query 134 ASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIV 193
+ T +YP++ +TRL ++ G+ DC KK+ R+ G + Y+G+ ++ GII
Sbjct 302 TAQTAIYPMEVMKTRLTLR----KTGQYSGMFDCAKKILRKEGVKAFYKGYVPNILGIIP 357
Query 194 YRGAYFGLYDA 204
Y G +Y+
Sbjct 358 YAGIDLAVYET 368
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/172 (20%), Positives = 80/172 (46%), Gaps = 10/172 (5%)
Query 13 IDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCK 72
+ + + F+AG ++ ++T + P+E +K + + T +YSG+ +C ++ +
Sbjct 287 VQSHERFMAGSLAGATAQTAIYPMEVMKTRLTLR---------KTGQYSGMFDCAKKILR 337
Query 73 EQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAG 132
++GV + ++G + N++ P + +T K + + + L + G ++
Sbjct 338 KEGVKAFYKGYVPNILGIIPYAGIDLAVYETLKNTWLSHYAKDTANPGVLVLLGCGTISS 397
Query 133 AASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGF 184
YPL RTR+ + + E + + +KK+ ++ GF LY+G
Sbjct 398 TCGQLASYPLALIRTRMQA-MASMEGSEQVSMSKLVKKIMQKEGFFGLYRGI 448
Score = 34.7 bits (78), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 50/106 (47%), Gaps = 5/106 (4%)
Query 83 NLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLTVVYPL 142
+L +IRY+ D+ I ++ + E+ + + +GG+AGA S T PL
Sbjct 159 DLQQIIRYWKKSTV-LDIGDSLT-IPDEFTEEEKTTGMWWKQLAAGGVAGAVSRTGTAPL 216
Query 143 DFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSV 188
D + + K + + L + K++ + G SL++G GV+V
Sbjct 217 DRMKVFMQVHSSKTNK---ISLVNGFKQMIKEGGVASLWRGNGVNV 259
> xla:379548 slc25a24-b, MGC68982, apc1, scamc-1, scamc1-B; solute
carrier family 25 (mitochondrial carrier; phosphate carrier),
member 24; K14684 solute carrier family 25 (mitochondrial
phosphate transporter), member 23/24/25/41
Length=473
Score = 107 bits (267), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 64/191 (33%), Positives = 101/191 (52%), Gaps = 25/191 (13%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K +AGG++ VS+T AP++R+K+++Q S S II ++ KE G+
Sbjct 196 KQLMAGGMAGAVSRTGTAPLDRLKVMMQVHGS---------KGNSNIITGLKQMVKEGGI 246
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFP----KYNQNEEFFKFFLTNVLSGGLAG 132
SLWRGN NVI+ P A F + +KK+F K E F ++G LAG
Sbjct 247 RSLWRGNGVNVIKIAPETAMKFWAYEQYKKLFTSESGKLGTAERF--------VAGSLAG 298
Query 133 AASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGII 192
A + T +YP++ +TRLA G ++ G+ DC KK+ ++ G + Y+G+ ++ GII
Sbjct 299 ATAQTSIYPMEVLKTRLAV----GRTGQYSGMFDCAKKIMQKEGIRAFYKGYIPNILGII 354
Query 193 VYRGAYFGLYD 203
Y G +Y+
Sbjct 355 PYAGIDLAIYE 365
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/173 (22%), Positives = 81/173 (46%), Gaps = 10/173 (5%)
Query 13 IDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCK 72
+ A+ F+AG ++ ++T + P+E +K + G T +YSG+ +C ++ +
Sbjct 285 LGTAERFVAGSLAGATAQTSIYPMEVLKTRLAV---------GRTGQYSGMFDCAKKIMQ 335
Query 73 EQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAG 132
++G+ + ++G + N++ P + +T K + + + + L + G +
Sbjct 336 KEGIRAFYKGYIPNILGIIPYAGIDLAIYETLKNYWLQNHAKDSANPGVLVLLGCGTASS 395
Query 133 AASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFG 185
YPL RTR+ + A + L +G +K+ + GFL LY+G G
Sbjct 396 TCGQLASYPLALIRTRMQAQASIEGAPQ-LNMGGLFRKIVAKEGFLGLYRGIG 447
> mmu:229731 Slc25a24, 2610016M12Rik; solute carrier family 25
(mitochondrial carrier, phosphate carrier), member 24; K14684
solute carrier family 25 (mitochondrial phosphate transporter),
member 23/24/25/41
Length=475
Score = 106 bits (265), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 61/187 (32%), Positives = 100/187 (53%), Gaps = 18/187 (9%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ +AGGV+ VS+T AP++R+K+++Q H + I F ++ KE G+
Sbjct 196 RQLLAGGVAGAVSRTSTAPLDRLKVMMQV----------HGSKSMNIFGGFRQMVKEGGI 245
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
SLWRGN NVI+ P A F + +KK+ + Q F+ F +SG +AGA +
Sbjct 246 RSLWRGNGTNVIKIAPETAVKFWAYEQYKKLLTEEGQKLGTFERF----ISGSMAGATAQ 301
Query 137 TVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRG 196
T +YP++ +TRLA ++ G+ C KK+ + GF + Y+G+ ++ GII Y G
Sbjct 302 TFIYPMEVLKTRLAV----AKTGQYSGIYGCAKKILKHEGFGAFYKGYIPNLLGIIPYAG 357
Query 197 AYFGLYD 203
+Y+
Sbjct 358 IDLAVYE 364
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/87 (20%), Positives = 39/87 (44%), Gaps = 6/87 (6%)
Query 22 GGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVYSLWR 81
G +S+ + P+ V+ +Q Q ++ P+ S ++ F R+ ++GV L+R
Sbjct 390 GALSSTCGQLASYPLALVRTRMQAQATVEG-----APQLS-MVGLFQRIVSKEGVSGLYR 443
Query 82 GNLANVIRYFPTQAFNFTFKDTFKKIF 108
G N ++ P ++ + K+
Sbjct 444 GITPNFMKVLPAVGISYVVYENMKQTL 470
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/121 (23%), Positives = 55/121 (45%), Gaps = 7/121 (5%)
Query 83 NLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLTVVYPL 142
++ +IR++ + D+ I ++ ++E+ + +L+GG+AGA S T PL
Sbjct 158 DIEEIIRFW-KHSTGIDIGDSLT-IPDEFTEDEKKSGQWWRQLLAGGVAGAVSRTSTAPL 215
Query 143 DFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRGAYFGLY 202
D R ++ V + G +++ + G SL++G G +V I F Y
Sbjct 216 D--RLKVMMQVHGSKSMNIFG---GFRQMVKEGGIRSLWRGNGTNVIKIAPETAVKFWAY 270
Query 203 D 203
+
Sbjct 271 E 271
> dre:569227 slc25a25b, scamc2b, si:dkey-7o20.1; solute carrier
family 25 (mitochondrial carrier; phosphate carrier), member
25b; K14684 solute carrier family 25 (mitochondrial phosphate
transporter), member 23/24/25/41
Length=469
Score = 102 bits (255), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 60/188 (31%), Positives = 103/188 (54%), Gaps = 16/188 (8%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
++ +AGG + VS+T AP++R+K+L+Q + A R GI F+++ +E G+
Sbjct 188 RHLVAGGGAGAVSRTCTAPLDRLKVLMQ----VHATRSNSM----GIAGGFTQMIREGGL 239
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
SLWRGN NV++ P A F + K++ N+E L ++SG LAGA +
Sbjct 240 RSLWRGNGINVLKIAPESAIKFMAYEQIKRLI---GSNQETLG-ILERLVSGSLAGAIAQ 295
Query 137 TVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRG 196
+ +YP++ +TRLA G ++ G+ DC K + ++ G + Y+G+ ++ GII Y G
Sbjct 296 SSIYPMEVLKTRLA----LGRTGQYSGIADCAKHIFKKEGMTAFYKGYIPNMLGIIPYAG 351
Query 197 AYFGLYDA 204
+Y+
Sbjct 352 IDLAVYET 359
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 36/188 (19%), Positives = 83/188 (44%), Gaps = 18/188 (9%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFS 68
N + + ++G ++ ++++ + P+E +K + G T +YSGI +C
Sbjct 274 NQETLGILERLVSGSLAGAIAQSSIYPMEVLKTRLAL---------GRTGQYSGIADCAK 324
Query 69 RVCKEQGVYSLWRGNLANVIRYFPTQAFNF----TFKDTFKKIFPKYNQNEEFFKFFLTN 124
+ K++G+ + ++G + N++ P + T K+++ + F + + F
Sbjct 325 HIFKKEGMTAFYKGYIPNMLGIIPYAGIDLAVYETLKNSWLQRFATDSADPGVFVLLACG 384
Query 125 VLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGF 184
+S AS YPL RTR+ + + + + + + + + R G + LY+G
Sbjct 385 TMSSTCGQLAS----YPLALVRTRMQAQASQEGSPQ-MTMSGLFRHIVRTEGAIGLYRGL 439
Query 185 GVSVQGII 192
+ +I
Sbjct 440 APNFMKVI 447
> ath:AT4G01100 ADNT1; ADNT1 (ADENINE NUCLEOTIDE TRANSPORTER 1);
ADP transmembrane transporter/ AMP transmembrane transporter/
ATP transmembrane transporter/ binding; K14684 solute carrier
family 25 (mitochondrial phosphate transporter), member
23/24/25/41
Length=352
Score = 99.8 bits (247), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 61/192 (31%), Positives = 98/192 (51%), Gaps = 12/192 (6%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K+ AGGV+ GVS+T VAP+ER+K+L+Q Q+ H +YSG + + + +G+
Sbjct 40 KSLFAGGVAGGVSRTAVAPLERMKILLQVQNP-------HNIKYSGTVQGLKHIWRTEGL 92
Query 77 YSLWRGNLANVIRYFPTQAFN-FTFKDTFKKIFPKYNQ---NEEFFKFFLTNVLSGGLAG 132
L++GN N R P A F+++ I Y Q NE L + +G AG
Sbjct 93 RGLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENAQLTPLLRLGAGATAG 152
Query 133 AASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGII 192
+++ YP+D R RL S ++ G+ + V R G +LY+G+ SV G++
Sbjct 153 IIAMSATYPMDMVRGRLTVQTAN-SPYQYRGIAHALATVLREEGPRALYRGWLPSVIGVV 211
Query 193 VYRGAYFGLYDA 204
Y G F +Y++
Sbjct 212 PYVGLNFSVYES 223
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 54/185 (29%), Positives = 85/185 (45%), Gaps = 30/185 (16%)
Query 20 IAGGVSAGV---SKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ G +AG+ S T + R ++ +QT +S +Y GI + + V +E+G
Sbjct 145 LGAGATAGIIAMSATYPMDMVRGRLTVQTANS--------PYQYRGIAHALATVLREEGP 196
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYN-----QNEEFFKFFLTNVLSGGLA 131
+L+RG L +VI P NF+ ++ K K N +N E +T + G +A
Sbjct 197 RALYRGWLPSVIGVVPYVGLNFSVYESLKDWLVKENPYGLVENNEL--TVVTRLTCGAIA 254
Query 132 GAASLTVVYPLDFARTRL-------ASDVGKGSAR-----EFLGLGDCIKKVSRRTGFLS 179
G T+ YPLD R R+ AS + G R E+ G+ D +K R GF +
Sbjct 255 GTVGQTIAYPLDVIRRRMQMVGWKDASAIVTGEGRSTASLEYTGMVDAFRKTVRHEGFGA 314
Query 180 LYQGF 184
LY+G
Sbjct 315 LYKGL 319
Score = 33.1 bits (74), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 40/88 (45%), Gaps = 2/88 (2%)
Query 118 FKFFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGF 177
FK ++ +GG+AG S T V PL+ R ++ V ++ G +K + R G
Sbjct 35 FKSICKSLFAGGVAGGVSRTAVAPLE--RMKILLQVQNPHNIKYSGTVQGLKHIWRTEGL 92
Query 178 LSLYQGFGVSVQGIIVYRGAYFGLYDAA 205
L++G G + I+ F Y+ A
Sbjct 93 RGLFKGNGTNCARIVPNSAVKFFSYEQA 120
> ath:AT4G26180 mitochondrial substrate carrier family protein
Length=325
Score = 99.0 bits (245), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 65/208 (31%), Positives = 108/208 (51%), Gaps = 26/208 (12%)
Query 8 DNNPLIDA----AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGI 63
+ N +ID+ AK IAGGV+ G++KT VAP+ER+K+L QT+ RD + G+
Sbjct 6 EKNGIIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQTR------RDEF--KRIGL 57
Query 64 INCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKI----FPKYNQNEEFFK 119
+ +++ K +G+ +RGN A+V R P A ++ + +++ FP +
Sbjct 58 VGSINKIGKTEGLMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGP---- 113
Query 120 FFLTNVLSGGLAGAASLTVVYPLDFARTRLA--SDVGKGSAREFL--GLGDCIKKVSRRT 175
L ++++G AG ++ YPLD RT+LA + V + + G+ DC + R +
Sbjct 114 --LLDLVAGSFAGGTAVLFTYPLDLVRTKLAYQTQVKAIPVEQIIYRGIVDCFSRTYRES 171
Query 176 GFLSLYQGFGVSVQGIIVYRGAYFGLYD 203
G LY+G S+ GI Y G F Y+
Sbjct 172 GARGLYRGVAPSLYGIFPYAGLKFYFYE 199
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 53/200 (26%), Positives = 89/200 (44%), Gaps = 18/200 (9%)
Query 9 NNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFS 68
PL+D +AG + G + P++ V+ + Q + AI Y GI++CFS
Sbjct 111 RGPLLD----LVAGSFAGGTAVLFTYPLDLVRTKLAYQTQVKAIPVEQII-YRGIVDCFS 165
Query 69 RVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSG 128
R +E G L+RG ++ FP F F + K+ P ++ + K ++ G
Sbjct 166 RTYRESGARGLYRGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLK-----LVCG 220
Query 129 GLAGAASLTVVYPLDFAR-----TRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQG 183
+AG T+ YPLD R RL S V + + R G + K++R G+ L+ G
Sbjct 221 SVAGLLGQTLTYPLDVVRRQMQVERLYSAVKEETRR---GTMQTLFKIAREEGWKQLFSG 277
Query 184 FGVSVQGIIVYRGAYFGLYD 203
++ ++ F +YD
Sbjct 278 LSINYLKVVPSVAIGFTVYD 297
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 50/105 (47%), Gaps = 5/105 (4%)
Query 3 EEKRRDNNPLI--DAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRY 60
EE +R P D + + G V+ + +T+ P++ V+ +Q + A+++ T R
Sbjct 199 EEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLDVVRRQMQVERLYSAVKE-ETRR- 256
Query 61 SGIINCFSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFK 105
G + ++ +E+G L+ G N ++ P+ A FT D K
Sbjct 257 -GTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFTVYDIMK 300
> ath:AT3G55640 mitochondrial substrate carrier family protein
Length=332
Score = 97.8 bits (242), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 54/189 (28%), Positives = 92/189 (48%), Gaps = 10/189 (5%)
Query 7 RDNNPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINC 66
+D I++A +AGG++ SKT AP+ R+ +L Q Q + R I++
Sbjct 26 QDQRSHIESASQLLAGGLAGAFSKTCTAPLSRLTILFQVQG---MHTNAAALRKPSILHE 82
Query 67 FSRVCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKK----IFPKYNQNEEFFKFFL 122
SR+ E+G+ + W+GNL + P + NF + +KK + N E
Sbjct 83 ASRILNEEGLKAFWKGNLVTIAHRLPYSSVNFYAYEHYKKFMYMVTGMENHKEGISSNLF 142
Query 123 TNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQ 182
+ ++GGLAG + + YPLD RTRLA+ + G+ ++ ++ G L LY+
Sbjct 143 VHFVAGGLAGITAASATYPLDLVRTRLAAQT---KVIYYSGIWHTLRSITTDEGILGLYK 199
Query 183 GFGVSVQGI 191
G G ++ G+
Sbjct 200 GLGTTLVGV 208
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/168 (22%), Positives = 78/168 (46%), Gaps = 12/168 (7%)
Query 18 NFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGVY 77
+F+AGG++ + + P++ V+ + Q + YSGI + + ++G+
Sbjct 144 HFVAGGLAGITAASATYPLDLVRTRLAAQTKVI--------YYSGIWHTLRSITTDEGIL 195
Query 78 SLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASLT 137
L++G ++ P+ A +F+ ++ + + ++ + ++ G L+G AS T
Sbjct 196 GLYKGLGTTLVGVGPSIAISFSVYESLRSYWRSTRPHDSPI---MVSLACGSLSGIASST 252
Query 138 VVYPLDFARTRLASDVGKGSAREF-LGLGDCIKKVSRRTGFLSLYQGF 184
+PLD R R + G A + GL +K++ + G LY+G
Sbjct 253 ATFPLDLVRRRKQLEGIGGRAVVYKTGLLGTLKRIVQTEGARGLYRGI 300
> hsa:114789 SLC25A25, KIAA1896, MCSC, MGC105138, MGC119514, MGC119515,
MGC119516, MGC119517, PCSCL, RP11-395P17.4, SCAMC-2;
solute carrier family 25 (mitochondrial carrier; phosphate
carrier), member 25; K14684 solute carrier family 25 (mitochondrial
phosphate transporter), member 23/24/25/41
Length=503
Score = 96.7 bits (239), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 55/189 (29%), Positives = 102/189 (53%), Gaps = 20/189 (10%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYS--GIINCFSRVCKEQ 74
++ +AGG + VS+T AP++R+K+L+Q H R + GI+ F+++ +E
Sbjct 222 RHLVAGGGAGAVSRTCTAPLDRLKVLMQV----------HASRSNNMGIVGGFTQMIREG 271
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G SLWRGN NV++ P A F + K++ ++E + +++G LAGA
Sbjct 272 GARSLWRGNGINVLKIAPESAIKFMAYEQIKRLV---GSDQETLRIH-ERLVAGSLAGAI 327
Query 135 SLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVY 194
+ + +YP++ +TR+A ++ G+ DC +++ R G + Y+G+ ++ GII Y
Sbjct 328 AQSSIYPMEVLKTRMALR----KTGQYSGMLDCARRILAREGVAAFYKGYVPNMLGIIPY 383
Query 195 RGAYFGLYD 203
G +Y+
Sbjct 384 AGIDLAVYE 392
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/176 (20%), Positives = 77/176 (43%), Gaps = 10/176 (5%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ +AG ++ ++++ + P+E +K + A+R T +YSG+++C R+ +GV
Sbjct 316 ERLVAGSLAGAIAQSSIYPMEVLKTRM-------ALRK--TGQYSGMLDCARRILAREGV 366
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
+ ++G + N++ P + +T K + ++ + G ++
Sbjct 367 AAFYKGYVPNMLGIIPYAGIDLAVYETLKNAWLQHYAVNSADPGVFVLLACGTMSSTCGQ 426
Query 137 TVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGII 192
YPL RTR+ + A E + + K + R G LY+G + +I
Sbjct 427 LASYPLALVRTRMQAQASIEGAPE-VTMSSLFKHILRTEGAFGLYRGLAPNFMKVI 481
> mmu:74686 4930443G12Rik; RIKEN cDNA 4930443G12 gene; K14684
solute carrier family 25 (mitochondrial phosphate transporter),
member 23/24/25/41
Length=473
Score = 94.0 bits (232), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 56/188 (29%), Positives = 101/188 (53%), Gaps = 18/188 (9%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K +A G+++ +++T AP+ER+K+ +Q Q S+ + G+++ F ++ KE G
Sbjct 196 KRLVAAGIASAITRTCTAPLERLKVTMQVQ-SLKVNK-------MGLVHMFKQMVKEGGF 247
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
+SLWRGN N+++ P A + +KK+ + + F +G +AGA S
Sbjct 248 FSLWRGNGVNILKIAPETAIKIGAYEQYKKLLSFDGDHLGVLQRF----TAGCMAGATSQ 303
Query 137 TVVYPLDFARTRL-ASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYR 195
T VYP++ +TRL S G E+ GL DC++K+ +R G + +G+ ++ II Y
Sbjct 304 TCVYPMEVIKTRLNLSKTG-----EYSGLVDCVRKLLKREGIQAFSKGYVPNLLSIIPYA 358
Query 196 GAYFGLYD 203
G +++
Sbjct 359 GLDLTIFE 366
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/185 (19%), Positives = 81/185 (43%), Gaps = 30/185 (16%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ F AG ++ S+T V P+E +K + T YSG+++C ++ K +G+
Sbjct 290 QRFTAGCMAGATSQTCVYPMEVIKTRLNL---------SKTGEYSGLVDCVRKLLKREGI 340
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
+ +G + N++ P + T + K + + + N ++ G+A
Sbjct 341 QAFSKGYVPNLLSIIPYAGLDLTIFELLKN---------HWLEHYAGNSVNPGIAIVLGC 391
Query 137 TVV---------YPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVS 187
+ V +PL RTR+ + + + +E + + I+++ + G ++GF +
Sbjct 392 STVSHTCGQLASFPLILVRTRMQAVMLE---KETVRMMQLIQEIYTKEGKKGFFRGFTPN 448
Query 188 VQGII 192
V ++
Sbjct 449 VLKLL 453
> ath:AT2G37890 mitochondrial substrate carrier family protein
Length=337
Score = 93.6 bits (231), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 57/182 (31%), Positives = 89/182 (48%), Gaps = 16/182 (8%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+N +AGG++ +SKT AP+ R+ +L Q Q +G + SR+ E+G
Sbjct 43 QNLLAGGIAGAISKTCTAPLARLTILFQLQG---MQSEGAVLSRPNLRREASRIINEEGY 99
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFF-------LTNVLSGG 129
+ W+GNL V+ P A NF + + F N N F + + +SGG
Sbjct 100 RAFWKGNLVTVVHRIPYTAVNFYAYEKYNLFF---NSNPVVQSFIGNTSGNPIVHFVSGG 156
Query 130 LAGAASLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQ 189
LAG + T YPLD RTRLA+ + +A + G+ + + R G L LY+G G ++
Sbjct 157 LAGITAATATYPLDLVRTRLAA---QRNAIYYQGIEHTFRTICREEGILGLYKGLGATLL 213
Query 190 GI 191
G+
Sbjct 214 GV 215
Score = 82.8 bits (203), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 57/196 (29%), Positives = 98/196 (50%), Gaps = 16/196 (8%)
Query 10 NPLIDAAKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSR 69
NP++ +F++GG++ + T P++ V+ + Q + AI Y GI + F
Sbjct 147 NPIV----HFVSGGLAGITAATATYPLDLVRTRLAAQRN--AIY------YQGIEHTFRT 194
Query 70 VCKEQGVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGG 129
+C+E+G+ L++G A ++ P+ A NF ++ K + + N+ + +++SGG
Sbjct 195 ICREEGILGLYKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPNDSDL---VVSLVSGG 251
Query 130 LAGAASLTVVYPLDFARTRLASDVGKGSAREF-LGLGDCIKKVSRRTGFLSLYQGFGVSV 188
LAGA S T YPLD R R+ + G AR + GL K + + GF +Y+G
Sbjct 252 LAGAVSSTATYPLDLVRRRMQVEGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEY 311
Query 189 QGIIVYRGAYFGLYDA 204
++ G F YDA
Sbjct 312 YKVVPGVGIVFMTYDA 327
> dre:406541 slc25a25a, slc25a, slc25a25, wu:fb13d12, wu:fd14e03,
zgc:77454; solute carrier family 25 (mitochondrial carrier;
phosphate carrier), member 25a; K14684 solute carrier family
25 (mitochondrial phosphate transporter), member 23/24/25/41
Length=469
Score = 93.2 bits (230), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 53/188 (28%), Positives = 97/188 (51%), Gaps = 16/188 (8%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
++ ++GG + VS+T AP++R+K+L+Q G + +++ +++ KE GV
Sbjct 188 RHLVSGGGAGAVSRTCTAPLDRLKVLMQVH--------GCQGKSMCLMSGLTQMIKEGGV 239
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
SLWRGN NVI+ P A F + K++ + + F ++G LAG +
Sbjct 240 RSLWRGNGINVIKIAPETALKFMAYEQIKRVMGSSQETLGISERF----VAGSLAGVIAQ 295
Query 137 TVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRG 196
+ +YP++ +TRLA ++ G+ DC K + + G + Y+G+ ++ GII Y G
Sbjct 296 STIYPMEVLKTRLALR----KTGQYKGISDCAKHILKTEGMSAFYKGYVPNMLGIIPYAG 351
Query 197 AYFGLYDA 204
+Y+
Sbjct 352 IDLAVYET 359
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/174 (22%), Positives = 82/174 (47%), Gaps = 20/174 (11%)
Query 16 AKNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQG 75
++ F+AG ++ ++++ + P+E +K + A+R T +Y GI +C + K +G
Sbjct 281 SERFVAGSLAGVIAQSTIYPMEVLKTRL-------ALRK--TGQYKGISDCAKHILKTEG 331
Query 76 VYSLWRGNLANVIRYFPTQAFNF----TFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLA 131
+ + ++G + N++ P + T K+T+ + + N + F +S
Sbjct 332 MSAFYKGYVPNMLGIIPYAGIDLAVYETLKNTWLQRYGTENADPGVFVLLACGTVSSTCG 391
Query 132 GAASLTVVYPLDFARTRLASDVG-KGSAREFLGLGDCIKKVSRRTGFLSLYQGF 184
AS YPL RTR+ + +GS++ + + K++ + G LY+G
Sbjct 392 QLAS----YPLALIRTRMQAQASVEGSSQ--VSMTGLFKQIMKTEGPTGLYRGL 439
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 43/95 (45%), Gaps = 5/95 (5%)
Query 110 KYNQNEEFFKFFLTNVLSGGLAGAASLTVVYPLDFARTRLASDVGKGSAREFL-GLGDCI 168
++ E + +++SGG AGA S T PLD + + +G + + GL I
Sbjct 175 EFTVEEHLTGMWWRHLVSGGGAGAVSRTCTAPLDRLKVLMQVHGCQGKSMCLMSGLTQMI 234
Query 169 KKVSRRTGFLSLYQGFGVSVQGIIVYRGAYFGLYD 203
K+ G SL++G G++V I F Y+
Sbjct 235 KE----GGVRSLWRGNGINVIKIAPETALKFMAYE 265
> mmu:227731 Slc25a25, 1110030N17Rik, MCSC, MGC36388, mKIAA1896;
solute carrier family 25 (mitochondrial carrier, phosphate
carrier), member 25; K14684 solute carrier family 25 (mitochondrial
phosphate transporter), member 23/24/25/41
Length=502
Score = 92.8 bits (229), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 53/189 (28%), Positives = 101/189 (53%), Gaps = 20/189 (10%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSG--IINCFSRVCKEQ 74
++ +AGG + VS+T AP++R+K+L+Q H R + I+ F+++ +E
Sbjct 221 RHLVAGGGAGAVSRTCTAPLDRLKVLMQV----------HASRSNNMCIVGGFTQMIREG 270
Query 75 GVYSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAA 134
G SLWRGN NV++ P A F + K++ ++E + +++G LAGA
Sbjct 271 GAKSLWRGNGINVLKIAPESAIKFMAYEQMKRLV---GSDQETLRIH-ERLVAGSLAGAI 326
Query 135 SLTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVY 194
+ + +YP++ +TR+A ++ G+ DC +++ + G + Y+G+ ++ GII Y
Sbjct 327 AQSSIYPMEVLKTRMALR----KTGQYSGMLDCARRILAKEGVAAFYKGYIPNMLGIIPY 382
Query 195 RGAYFGLYD 203
G +Y+
Sbjct 383 AGIDLAVYE 391
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 39/177 (22%), Positives = 82/177 (46%), Gaps = 12/177 (6%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
+ +AG ++ ++++ + P+E +K + A+R T +YSG+++C R+ ++GV
Sbjct 315 ERLVAGSLAGAIAQSSIYPMEVLKTRM-------ALR--KTGQYSGMLDCARRILAKEGV 365
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIF-PKYNQNEEFFKFFLTNVLSGGLAGAAS 135
+ ++G + N++ P + +T K + +Y N F+ + G ++
Sbjct 366 AAFYKGYIPNMLGIIPYAGIDLAVYETLKNTWLQRYAVNSADPGVFVL-LACGTISSTCG 424
Query 136 LTVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGII 192
YPL RTR+ + A E + + K++ R G LY+G + +I
Sbjct 425 QLASYPLALVRTRMQAQASIEGAPE-VTMSSLFKQILRTEGAFGLYRGLAPNFMKVI 480
> mmu:66972 Slc25a23, 2310067G05Rik; solute carrier family 25
(mitochondrial carrier; phosphate carrier), member 23; K14684
solute carrier family 25 (mitochondrial phosphate transporter),
member 23/24/25/41
Length=467
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 57/187 (30%), Positives = 92/187 (49%), Gaps = 16/187 (8%)
Query 17 KNFIAGGVSAGVSKTIVAPVERVKMLIQTQDSIPAIRDGHTPRYSGIINCFSRVCKEQGV 76
K +AG V+ VS+T AP++R+K+ +Q S I+ + +E GV
Sbjct 186 KQLVAGAVAGAVSRTGTAPLDRLKVFMQVHAS--------KSNRLNILGGLRNMIQEGGV 237
Query 77 YSLWRGNLANVIRYFPTQAFNFTFKDTFKKIFPKYNQNEEFFKFFLTNVLSGGLAGAASL 136
SLWRGN NV++ P A F + K+ + + F ++G LAGA +
Sbjct 238 LSLWRGNGINVLKIAPESAIKFMAYEQIKRAIRGQQETLHVQERF----VAGSLAGATAQ 293
Query 137 TVVYPLDFARTRLASDVGKGSAREFLGLGDCIKKVSRRTGFLSLYQGFGVSVQGIIVYRG 196
T++YP++ +TRL ++ GL DC K++ R G + Y+G+ +V GII Y G
Sbjct 294 TIIYPMEVLKTRLTLR----RTGQYKGLLDCAKRILEREGPRAFYRGYLPNVLGIIPYAG 349
Query 197 AYFGLYD 203
+Y+
Sbjct 350 IDLAVYE 356
Lambda K H
0.323 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6623499460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40