bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1177_orf1
Length=161
Score E
Sequences producing significant alignments: (Bits) Value
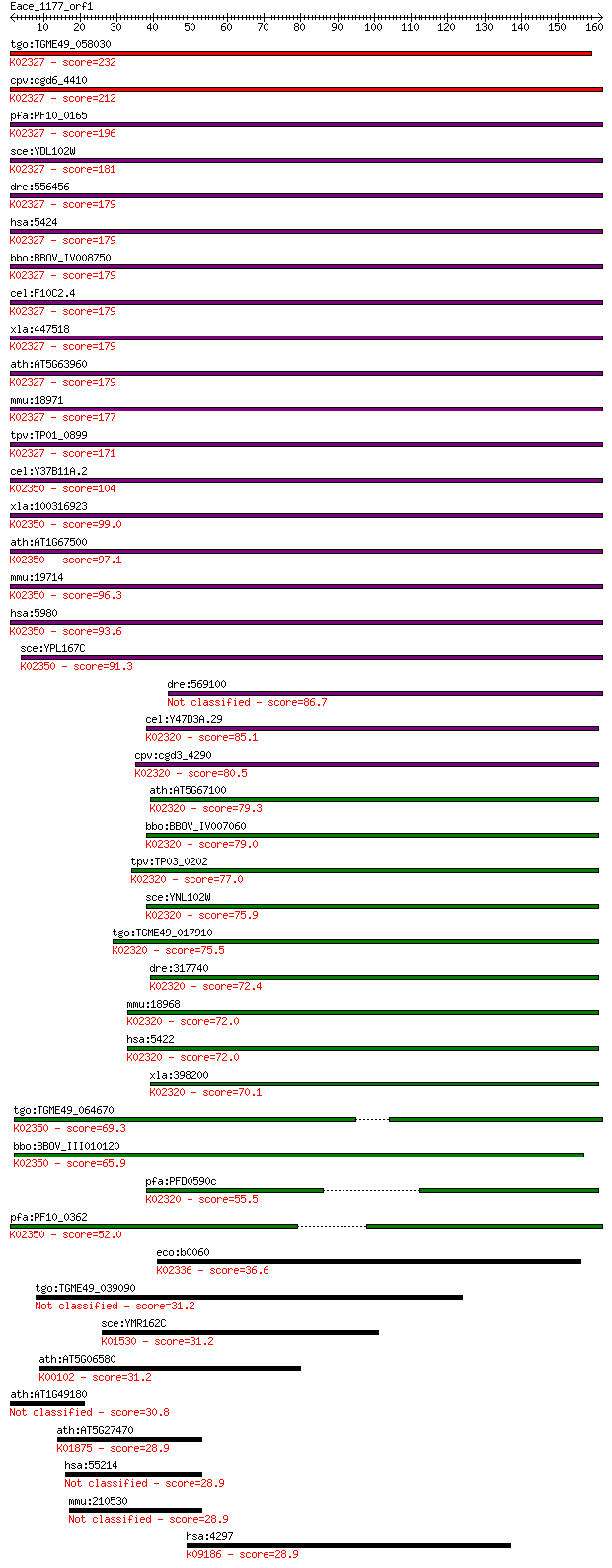
tgo:TGME49_058030 DNA polymerase delta catalytic subunit, puta... 232 5e-61
cpv:cgd6_4410 DNA polymerase delta catalytic subunit. DNAQ-lik... 212 3e-55
pfa:PF10_0165 DNA polymerase delta catalytic subunit; K02327 D... 196 3e-50
sce:YDL102W POL3, CDC2, HPR6, TEX1; Catalytic subunit of DNA p... 181 1e-45
dre:556456 pold1, MGC152688, fla, im:7139126, zgc:152688; poly... 179 3e-45
hsa:5424 POLD1, CDC2, POLD; polymerase (DNA directed), delta 1... 179 3e-45
bbo:BBOV_IV008750 23.m05784; DNA polymerase delta subunit (EC:... 179 3e-45
cel:F10C2.4 hypothetical protein; K02327 DNA polymerase delta ... 179 4e-45
xla:447518 pold1, MGC83185, POL3, cdc2, pold; polymerase (DNA ... 179 5e-45
ath:AT5G63960 EMB2780 (EMBRYO DEFECTIVE 2780); DNA binding / D... 179 5e-45
mmu:18971 Pold1, 125kDa; polymerase (DNA directed), delta 1, c... 177 2e-44
tpv:TP01_0899 DNA polymerase delta catalytic subunit; K02327 D... 171 1e-42
cel:Y37B11A.2 hypothetical protein; K02350 DNA polymerase zeta... 104 1e-22
xla:100316923 rev3l; REV3-like, catalytic subunit of DNA polym... 99.0 7e-21
ath:AT1G67500 ATREV3; ATREV3 (ARABIDOPSIS THALIANA RECOVERY PR... 97.1 2e-20
mmu:19714 Rev3l, Rev, Rev3, Sez4; REV3-like, catalytic subunit... 96.3 4e-20
hsa:5980 REV3L, POLZ, REV3; REV3-like, catalytic subunit of DN... 93.6 3e-19
sce:YPL167C REV3, PSO1; Catalytic subunit of DNA polymerase ze... 91.3 1e-18
dre:569100 rev3l, im:7037179, wu:fb98b12, wu:fu65g10; REV3-lik... 86.7 3e-17
cel:Y47D3A.29 hypothetical protein; K02320 DNA polymerase alph... 85.1 8e-17
cpv:cgd3_4290 DNA polymerase alpha catalytic subunit ; K02320 ... 80.5 2e-15
ath:AT5G67100 ICU2; ICU2 (INCURVATA2); DNA-directed DNA polyme... 79.3 4e-15
bbo:BBOV_IV007060 23.m06311; DNA polymerase alpha subunit (EC:... 79.0 6e-15
tpv:TP03_0202 DNA polymerase alpha (EC:2.7.7.7); K02320 DNA po... 77.0 2e-14
sce:YNL102W POL1, CDC17, CRT5, HPR3; Catalytic subunit of the ... 75.9 6e-14
tgo:TGME49_017910 DNA polymerase alpha catalytic subunit (EC:2... 75.5 8e-14
dre:317740 pola1, cb678, pola; polymerase (DNA directed), alph... 72.4 6e-13
mmu:18968 Pola1, AW321876, Pola; polymerase (DNA directed), al... 72.0 7e-13
hsa:5422 POLA1, DKFZp686K1672, POLA, p180; polymerase (DNA dir... 72.0 9e-13
xla:398200 pola1, pola; polymerase (DNA directed), alpha 1, ca... 70.1 3e-12
tgo:TGME49_064670 DNA polymerase zeta catalytic subunit, putat... 69.3 6e-12
bbo:BBOV_III010120 17.m07879; DNA polymerase family B family p... 65.9 6e-11
pfa:PFD0590c DNA polymerase alpha; K02320 DNA polymerase alpha... 55.5 7e-08
pfa:PF10_0362 DNA polymerase zeta catalytic subunit, putative;... 52.0 8e-07
eco:b0060 polB, dinA, ECK0061, JW0059; DNA polymerase II (EC:2... 36.6 0.039
tgo:TGME49_039090 surface antigen, putative 31.2 1.7
sce:YMR162C DNF3; Aminophospholipid translocase (flippase) tha... 31.2 1.7
ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4); ... 31.2 1.7
ath:AT1G49180 protein kinase family protein 30.8 2.2
ath:AT5G27470 seryl-tRNA synthetase / serine--tRNA ligase (EC:... 28.9 6.6
hsa:55214 LEPREL1, FLJ10718, MLAT4, P3H2; leprecan-like 1 (EC:... 28.9 6.7
mmu:210530 Leprel1, 4832416N06, AW553532, Mlat4, P3H2; lepreca... 28.9 7.8
hsa:4297 MLL, ALL-1, CXXC7, FLJ11783, HRX, HTRX1, KMT2A, MLL/G... 28.9 7.8
> tgo:TGME49_058030 DNA polymerase delta catalytic subunit, putative
(EC:2.7.7.7); K02327 DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1268
Score = 232 bits (591), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 112/159 (70%), Positives = 132/159 (83%), Gaps = 1/159 (0%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LLTRGQQIKVT+QLLRKC++L+Y+VP VKR GGD +YEGATVLEP KGFY+ PI+TLD
Sbjct 712 LLTRGQQIKVTAQLLRKCKELNYVVPVVKRTGGDNSVQYEGATVLEPRKGFYDKPIATLD 771
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQ-DTLTHTTSSPRHTFVKASVRRGVLPMIVE 119
FASLYPSIM+AHNICYSTLV + T+ D +T TT+SP H FVK +RRGVLPMIVE
Sbjct 772 FASLYPSIMIAHNICYSTLVASSAAHTMNNPDDVTVTTTSPPHKFVKKHIRRGVLPMIVE 831
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSV 158
ELIAARK A+ EM+ A+D+ + VLNGRQLALKI+ANSV
Sbjct 832 ELIAARKAARKEMAAAKDEMTRQVLNGRQLALKISANSV 870
> cpv:cgd6_4410 DNA polymerase delta catalytic subunit. DNAQ-like
3'-5' exonuclease; RNAseH fold ; K02327 DNA polymerase delta
subunit 1 [EC:2.7.7.7]
Length=1084
Score = 212 bits (540), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 105/161 (65%), Positives = 130/161 (80%), Gaps = 3/161 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL+RGQQIKVTSQ+LRKC+ ++L+P+VK D D++YEGATVLEPLKGFY PISTLD
Sbjct 531 LLSRGQQIKVTSQILRKCKSTNFLMPTVKN-QSDGDNQYEGATVLEPLKGFYKDPISTLD 589
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEE 120
FASLYPSIM+AHNICYSTL+ +++ + +D TS H+FV +SVR+G+LP+IVEE
Sbjct 590 FASLYPSIMIAHNICYSTLIPPLNLKNVPED--LRKTSPTGHSFVLSSVRKGILPLIVEE 647
Query 121 LIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
LIAARK AK EM +A D + S+LNGRQLALK +ANSVYGY
Sbjct 648 LIAARKKAKKEMEEATDPTLISILNGRQLALKTSANSVYGY 688
> pfa:PF10_0165 DNA polymerase delta catalytic subunit; K02327
DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1094
Score = 196 bits (498), Expect = 3e-50, Method: Composition-based stats.
Identities = 99/162 (61%), Positives = 124/162 (76%), Gaps = 2/162 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LLTRGQQIKVTSQL RKC++L+Y++PS + + ++KYEGATVLEP+KG+Y PISTLD
Sbjct 540 LLTRGQQIKVTSQLYRKCKELNYVIPST-YMKVNTNEKYEGATVLEPIKGYYIEPISTLD 598
Query 61 FASLYPSIMMAHNICYSTLVKGT-DVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVE 119
FASLYPSIM+AHN+CYSTL+K +V LQ D +T FVK +V++G+LP+IVE
Sbjct 599 FASLYPSIMIAHNLCYSTLIKSNHEVSDLQNDDITTIQGKNNLKFVKKNVKKGILPLIVE 658
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
ELI ARK K + ++ K VLNGRQLALKI+ANSVYGY
Sbjct 659 ELIEARKKVKLLIKNEKNNITKMVLNGRQLALKISANSVYGY 700
> sce:YDL102W POL3, CDC2, HPR6, TEX1; Catalytic subunit of DNA
polymerase delta; required for chromosomal DNA replication
during mitosis and meiosis, intragenic recombination, repair
of double strand DNA breaks, and DNA replication during nucleotide
excision repair (NER) (EC:2.7.7.7); K02327 DNA polymerase
delta subunit 1 [EC:2.7.7.7]
Length=1097
Score = 181 bits (459), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 87/162 (53%), Positives = 117/162 (72%), Gaps = 3/162 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL RGQQIKV SQL RKC ++ ++P+++ D D+YEGATV+EP++G+Y++PI+TLD
Sbjct 551 LLARGQQIKVVSQLFRKCLEIDTVIPNMQSQASD--DQYEGATVIEPIRGYYDVPIATLD 608
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPR-HTFVKASVRRGVLPMIVE 119
F SLYPSIMMAHN+CY+TL V+ L +P FV RRG+LP+I++
Sbjct 609 FNSLYPSIMMAHNLCYTTLCNKATVERLNLKIDEDYVITPNGDYFVTTKRRRGILPIILD 668
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
ELI+ARK AK ++ +D F + VLNGRQLALKI+ANSVYG+
Sbjct 669 ELISARKRAKKDLRDEKDPFKRDVLNGRQLALKISANSVYGF 710
> dre:556456 pold1, MGC152688, fla, im:7139126, zgc:152688; polymerase
(DNA directed), delta 1, catalytic subunit (EC:2.7.7.7);
K02327 DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1105
Score = 179 bits (455), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 90/161 (55%), Positives = 120/161 (74%), Gaps = 3/161 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL+RGQQIKV SQLLR+ + ++P VK GG+ Y GATV+EP KG+Y++PI+TLD
Sbjct 550 LLSRGQQIKVVSQLLRQAMKQDLVMPVVKTEGGE---DYTGATVIEPEKGYYSVPIATLD 606
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEE 120
F+SLYPSIMMAHN+CY+TL++ + ++ L + FVK+SVR+G+LP I+E
Sbjct 607 FSSLYPSIMMAHNLCYTTLLQKSQIEKLGLGPDDFIKTPTGDLFVKSSVRKGLLPEILEN 666
Query 121 LIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
L++ARK AKAE+ K D F K VL+GRQLALKI+ANSVYG+
Sbjct 667 LLSARKRAKAELKKETDPFKKQVLDGRQLALKISANSVYGF 707
> hsa:5424 POLD1, CDC2, POLD; polymerase (DNA directed), delta
1, catalytic subunit 125kDa (EC:2.7.7.7); K02327 DNA polymerase
delta subunit 1 [EC:2.7.7.7]
Length=1107
Score = 179 bits (455), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 91/163 (55%), Positives = 120/163 (73%), Gaps = 7/163 (4%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL+RGQQ+KV SQLLR+ L+P VK GG+ Y GATV+EPLKG+Y++PI+TLD
Sbjct 546 LLSRGQQVKVVSQLLRQAMHEGLLMPVVKSEGGE---DYTGATVIEPLKGYYDVPIATLD 602
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQT--LQQDTLTHTTSSPRHTFVKASVRRGVLPMIV 118
F+SLYPSIMMAHN+CY+TL++ Q L +D T + FVK SVR+G+LP I+
Sbjct 603 FSSLYPSIMMAHNLCYTTLLRPGTAQKLGLTEDQFIRTPTG--DEFVKTSVRKGLLPQIL 660
Query 119 EELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
E L++ARK AKAE++K D + VL+GRQLALK++ANSVYG+
Sbjct 661 ENLLSARKRAKAELAKETDPLRRQVLDGRQLALKVSANSVYGF 703
> bbo:BBOV_IV008750 23.m05784; DNA polymerase delta subunit (EC:2.7.7.7);
K02327 DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1065
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 90/163 (55%), Positives = 112/163 (68%), Gaps = 2/163 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRV--GGDRDDKYEGATVLEPLKGFYNMPIST 58
L++RGQQI+VT Q+ R+C+ + Y VPSV + + YEGATVLEP KG++ I+
Sbjct 508 LISRGQQIRVTMQIYRQCKLMGYAVPSVNAAQRAANSEASYEGATVLEPKKGYHRNAIAV 567
Query 59 LDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIV 118
LDF SLYPSIM+AHN+CYSTLV D + LT P F+KA VR+G+LP+IV
Sbjct 568 LDFQSLYPSIMIAHNLCYSTLVPPADASKYPAEDLTTVPGHPGLYFIKAHVRKGMLPLIV 627
Query 119 EELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
E LI ARK AK EM D ++SVL+GRQLALKIT NSVYGY
Sbjct 628 ERLIEARKKAKEEMKVCTDPMLRSVLDGRQLALKITTNSVYGY 670
> cel:F10C2.4 hypothetical protein; K02327 DNA polymerase delta
subunit 1 [EC:2.7.7.7]
Length=1081
Score = 179 bits (453), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 86/161 (53%), Positives = 120/161 (74%), Gaps = 5/161 (3%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LLT+GQQIK+ S +LR+C+Q ++ +P ++ GD + YEGATV++P++GFYN PI+TLD
Sbjct 516 LLTKGQQIKILSMMLRRCKQNNFFLPVIEANSGD-GEGYEGATVIDPIRGFYNEPIATLD 574
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEE 120
FASLYPSIM+AHN+CY+TL+K Q ++ + T S F S RRG+LP I+E+
Sbjct 575 FASLYPSIMIAHNLCYTTLLKSP--QGVENEDYIRTPSG--QYFATKSKRRGLLPEILED 630
Query 121 LIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
++AARK AK +M +D+F + V NGRQLALKI+ANSVYG+
Sbjct 631 ILAARKRAKNDMKNEKDEFKRMVYNGRQLALKISANSVYGF 671
> xla:447518 pold1, MGC83185, POL3, cdc2, pold; polymerase (DNA
directed), delta 1, catalytic subunit 125kDa (EC:2.7.7.7);
K02327 DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1109
Score = 179 bits (453), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 88/161 (54%), Positives = 118/161 (73%), Gaps = 3/161 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL+RGQQIKV SQLLR+ + ++P V+ GG+ Y GATV+EPLKG+Y++PI+TLD
Sbjct 551 LLSRGQQIKVVSQLLRQAMKQDLVMPVVRSEGGE---DYTGATVIEPLKGYYDVPIATLD 607
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEE 120
F+SLYPSIMMAHN+CY+TL++ V+ + + FVKASVR+G+LP I+E
Sbjct 608 FSSLYPSIMMAHNLCYTTLLQSGSVEKYGLNPEDFIKTPTGDCFVKASVRKGLLPEILEN 667
Query 121 LIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
L+ ARK AK E+ K D F + VL+GRQLALK++ANSVYG+
Sbjct 668 LLCARKRAKTELKKETDPFKQKVLDGRQLALKVSANSVYGF 708
> ath:AT5G63960 EMB2780 (EMBRYO DEFECTIVE 2780); DNA binding /
DNA-directed DNA polymerase/ nucleic acid binding / nucleotide
binding; K02327 DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1095
Score = 179 bits (453), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 93/162 (57%), Positives = 121/162 (74%), Gaps = 3/162 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL RGQ IKV SQLLRK +Q + ++P+ K+ G ++ YEGATVLE GFY PI+TLD
Sbjct 542 LLARGQSIKVLSQLLRKGKQKNLVLPNAKQSGSEQG-TYEGATVLEARTGFYEKPIATLD 600
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPR-HTFVKASVRRGVLPMIVE 119
FASLYPSIMMA+N+CY TLV DV+ L H T +P TFVK ++++G+LP I+E
Sbjct 601 FASLYPSIMMAYNLCYCTLVTPEDVRKLNLPP-EHVTKTPSGETFVKQTLQKGILPEILE 659
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
EL+ ARK AKA++ +A+D K+VL+GRQLALKI+ANSVYG+
Sbjct 660 ELLTARKRAKADLKEAKDPLEKAVLDGRQLALKISANSVYGF 701
> mmu:18971 Pold1, 125kDa; polymerase (DNA directed), delta 1,
catalytic subunit (EC:2.7.7.7); K02327 DNA polymerase delta
subunit 1 [EC:2.7.7.7]
Length=1105
Score = 177 bits (448), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 90/163 (55%), Positives = 121/163 (74%), Gaps = 7/163 (4%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LLTRGQQ+KV SQLLR+ + L+P VK G + Y GATV+EPLKG+Y++PI+TLD
Sbjct 544 LLTRGQQVKVVSQLLRQAMRQGLLMPVVKTEGSE---DYTGATVIEPLKGYYDVPIATLD 600
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQT--LQQDTLTHTTSSPRHTFVKASVRRGVLPMIV 118
F+SLYPSIMMAHN+CY+TL++ Q L+ D T + FVK+SVR+G+LP I+
Sbjct 601 FSSLYPSIMMAHNLCYTTLLRPGAAQKLGLKPDEFIKTPTG--DEFVKSSVRKGLLPQIL 658
Query 119 EELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
E L++ARK AKAE+++ D + VL+GRQLALK++ANSVYG+
Sbjct 659 ENLLSARKRAKAELAQETDPLRRQVLDGRQLALKVSANSVYGF 701
> tpv:TP01_0899 DNA polymerase delta catalytic subunit; K02327
DNA polymerase delta subunit 1 [EC:2.7.7.7]
Length=1095
Score = 171 bits (432), Expect = 1e-42, Method: Composition-based stats.
Identities = 81/163 (49%), Positives = 113/163 (69%), Gaps = 2/163 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVG--GDRDDKYEGATVLEPLKGFYNMPIST 58
L+TRGQQI+VT Q+ ++C++++Y++P + G ++ YEGATVLEPLKG++ PIS
Sbjct 532 LITRGQQIRVTMQIYKQCKKMNYVIPVIAGGGKSSSNENSYEGATVLEPLKGYHKNPISV 591
Query 59 LDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIV 118
LDF SLYPSIM+AHNICYSTL+ + + + + F+ R+G+LP+IV
Sbjct 592 LDFQSLYPSIMIAHNICYSTLLNYNTINNYKPEDIVTVPGYNDICFINVKKRKGILPIIV 651
Query 119 EELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
E LI RK AK M++ +D+ +K V +GRQLALKIT NSVYGY
Sbjct 652 ENLINERKKAKKMMNECKDEMLKKVYDGRQLALKITTNSVYGY 694
> cel:Y37B11A.2 hypothetical protein; K02350 DNA polymerase zeta
subunit [EC:2.7.7.7]
Length=1133
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 64/189 (33%), Positives = 103/189 (54%), Gaps = 28/189 (14%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEG-ATVLEPLKGFYNMPISTL 59
+ TRG Q++V S LLR +++++ PS+ + + E +LEP Y P+ L
Sbjct 578 VWTRGSQLRVESMLLRLAHRMNFVAPSITHLQRNMMGSPEQLQLILEPQSKVYFDPVIVL 637
Query 60 DFASLYPSIMMAHNICYST-------LVKGTDVQTLQQDTL------------------- 93
DF SLYPS+++A+N CYST LV+ D +++ +
Sbjct 638 DFQSLYPSMVIAYNYCYSTILGKIGNLVQMNDESRNREEIVLGAIKYHPSKDDIVKLVAY 697
Query 94 THTTSSPRHT-FVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALK 152
+SP + FVK S R GV+P+++ E++AAR + K+ M + ++K +K +L+ RQLALK
Sbjct 698 KEVCASPLASMFVKKSKREGVMPLLLREILAARIMVKSAMKRTKNKKLKRILDARQLALK 757
Query 153 ITANSVYGY 161
+ AN YGY
Sbjct 758 LVANVSYGY 766
> xla:100316923 rev3l; REV3-like, catalytic subunit of DNA polymerase
zeta; K02350 DNA polymerase zeta subunit [EC:2.7.7.7]
Length=3139
Score = 99.0 bits (245), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 99/182 (54%), Gaps = 22/182 (12%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHY--LVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPIST 58
+LTRG Q +V S +LR + ++Y L PS+++ R + V+EP FY+ I
Sbjct 2563 VLTRGSQYRVESMMLRIAKPMNYIPLTPSIQQRSQMRAPQC-IPLVMEPESRFYSNCILV 2621
Query 59 LDFASLYPSIMMAHNICYSTLVKGTD-----------VQTLQ--QDTL----THTTSSPR 101
LDF SLYPSI++A+N CYST + + +LQ D L T SP
Sbjct 2622 LDFQSLYPSIVIAYNYCYSTCLGNVENLGKYEDFKFGCSSLQVPPDLLYQLRNDITVSPN 2681
Query 102 H-TFVKASVRRGVLPMIVEELIAARKVAKAEMSK-AEDKFMKSVLNGRQLALKITANSVY 159
FVKA VR+GVLP ++EE++ R + K M +DK + +L+ RQL LK+ AN +
Sbjct 2682 GVAFVKACVRKGVLPRMLEEILKTRIMVKQSMKNYKQDKSLTRLLDARQLGLKLIANVTF 2741
Query 160 GY 161
GY
Sbjct 2742 GY 2743
> ath:AT1G67500 ATREV3; ATREV3 (ARABIDOPSIS THALIANA RECOVERY
PROTEIN 3); DNA binding / DNA-directed DNA polymerase; K02350
DNA polymerase zeta subunit [EC:2.7.7.7]
Length=1890
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 63/182 (34%), Positives = 94/182 (51%), Gaps = 25/182 (13%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGAT-----VLEPLKGFYNMP 55
+L+RG Q +V S LLR +YL S G++ + A V+EP FY+ P
Sbjct 1299 VLSRGSQYRVESMLLRLAHTQNYLAIS----PGNQQVASQPAMECVPLVMEPESAFYDDP 1354
Query 56 ISTLDFASLYPSIMMAHNICYSTLVK----------GTDVQTLQQDT---LTHTTSSPRH 102
+ LDF SLYPS+++A+N+C+ST + G +L D L +P
Sbjct 1355 VIVLDFQSLYPSMIIAYNLCFSTCLGKLAHLKMNTLGVSSYSLDLDVLQDLNQILQTPNS 1414
Query 103 T-FVKASVRRGVLPMIVEELIAARKVAKAEMSK--AEDKFMKSVLNGRQLALKITANSVY 159
+V VRRG+LP ++EE+++ R + K M K + + + N RQLALK+ AN Y
Sbjct 1415 VMYVPPEVRRGILPRLLEEILSTRIMVKKAMKKLTPSEAVLHRIFNARQLALKLIANVTY 1474
Query 160 GY 161
GY
Sbjct 1475 GY 1476
> mmu:19714 Rev3l, Rev, Rev3, Sez4; REV3-like, catalytic subunit
of DNA polymerase zeta RAD54 like (S. cerevisiae) (EC:2.7.7.7);
K02350 DNA polymerase zeta subunit [EC:2.7.7.7]
Length=3122
Score = 96.3 bits (238), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 65/182 (35%), Positives = 98/182 (53%), Gaps = 22/182 (12%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYL--VPSVKRVGGDRDDKYEGATVLEPLKGFYNMPIST 58
+LTRG Q +V S +LR + ++Y+ PS+++ R + ++EP FY+ +
Sbjct 2546 VLTRGSQYRVESMMLRIAKPMNYIPVTPSIQQRSQMRAPQC-VPLIMEPESRFYSNSVLV 2604
Query 59 LDFASLYPSIMMAHNICYSTLVK-------------GTDVQTLQQDTLTH----TTSSPR 101
LDF SLYPSI++A+N C+ST + G + D L T SP
Sbjct 2605 LDFQSLYPSIVIAYNYCFSTCLGHVENLGKYDEFKFGCTSLRVPPDLLYQIRHDVTVSPN 2664
Query 102 H-TFVKASVRRGVLPMIVEELIAARKVAKAEM-SKAEDKFMKSVLNGRQLALKITANSVY 159
FVK SVR+GVLP ++EE++ R + K M S +D+ + +LN RQL LK+ AN +
Sbjct 2665 GVAFVKPSVRKGVLPRMLEEILKTRLMVKQSMKSYKQDRALSRMLNARQLGLKLIANVTF 2724
Query 160 GY 161
GY
Sbjct 2725 GY 2726
> hsa:5980 REV3L, POLZ, REV3; REV3-like, catalytic subunit of
DNA polymerase zeta (yeast) (EC:2.7.7.7); K02350 DNA polymerase
zeta subunit [EC:2.7.7.7]
Length=3130
Score = 93.6 bits (231), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 64/182 (35%), Positives = 98/182 (53%), Gaps = 22/182 (12%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYL--VPSVKRVGGDRDDKYEGATVLEPLKGFYNMPIST 58
+LTRG Q +V S +LR + ++Y+ PSV++ R + ++EP FY+ +
Sbjct 2554 VLTRGSQYRVESMMLRIAKPMNYIPVTPSVQQRSQMRAPQC-VPLIMEPESRFYSNSVLV 2612
Query 59 LDFASLYPSIMMAHNICYSTLVK-------------GTDVQTLQQDTLTHT----TSSPR 101
LDF SLYPSI++A+N C+ST + G + D L T SP
Sbjct 2613 LDFQSLYPSIVIAYNYCFSTCLGHVENLGKYDEFKFGCTSLRVPPDLLYQVRHDITVSPN 2672
Query 102 H-TFVKASVRRGVLPMIVEELIAARKVAKAEM-SKAEDKFMKSVLNGRQLALKITANSVY 159
FVK SVR+GVLP ++EE++ R + K M + +D+ + +L+ RQL LK+ AN +
Sbjct 2673 GVAFVKPSVRKGVLPRMLEEILKTRFMVKQSMKAYKQDRALSRMLDARQLGLKLIANVTF 2732
Query 160 GY 161
GY
Sbjct 2733 GY 2734
> sce:YPL167C REV3, PSO1; Catalytic subunit of DNA polymerase
zeta, involved in translesion synthesis during post-replication
repair; required for mutagenesis induced by DNA damage;
involved in double-strand break repair (EC:2.7.7.7); K02350
DNA polymerase zeta subunit [EC:2.7.7.7]
Length=1504
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 61/182 (33%), Positives = 93/182 (51%), Gaps = 28/182 (15%)
Query 4 RGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGAT-----VLEPLKGFYNMPIST 58
RG Q KV S L+R C+ +++ S G +D + + A V+EP FY P+
Sbjct 918 RGSQFKVESFLIRICKSESFILLS----PGKKDVRKQKALECVPLVMEPESAFYKSPLIV 973
Query 59 LDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSS-PRH--------------- 102
LDF SLYPSIM+ +N CYST++ L ++ L + S PR+
Sbjct 974 LDFQSLYPSIMIGYNYCYSTMIGRVREINLTENNLGVSKFSLPRNILALLKNDVTIAPNG 1033
Query 103 -TFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDK--FMKSVLNGRQLALKITANSVY 159
+ K SVR+ L ++ +++ R + K M++ D +K +LN +QLALK+ AN Y
Sbjct 1034 VVYAKTSVRKSTLSKMLTDILDVRVMIKKTMNEIGDDNTTLKRLLNNKQLALKLLANVTY 1093
Query 160 GY 161
GY
Sbjct 1094 GY 1095
> dre:569100 rev3l, im:7037179, wu:fb98b12, wu:fu65g10; REV3-like,
catalytic subunit of DNA polymerase zeta (yeast)
Length=561
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 54/137 (39%), Positives = 74/137 (54%), Gaps = 19/137 (13%)
Query 44 VLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVK-------------GTDVQTLQQ 90
V+EP FY+ + LDF SLYPSI++A+N CYST + G +
Sbjct 25 VMEPESRFYSNSVIVLDFQSLYPSIVIAYNYCYSTCLGHIENLGTYDEFKFGCASLRVPP 84
Query 91 DTL----THTTSSPRH-TFVKASVRRGVLPMIVEELIAARKVAKAEM-SKAEDKFMKSVL 144
D L T SP FVK+SVRRGVLP ++EE++ R + K M + DK + +L
Sbjct 85 DLLYQLRNDITVSPNGVAFVKSSVRRGVLPRMLEEILKTRIMVKQSMKAYRNDKALSRLL 144
Query 145 NGRQLALKITANSVYGY 161
+ RQL LK+ AN +GY
Sbjct 145 DARQLGLKLIANVTFGY 161
> cel:Y47D3A.29 hypothetical protein; K02320 DNA polymerase alpha
subunit A [EC:2.7.7.7]
Length=1428
Score = 85.1 bits (209), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 49/123 (39%), Positives = 68/123 (55%), Gaps = 13/123 (10%)
Query 38 KYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTT 97
+Y G VLEP KG Y I LDF SLYPSI+ +NICY+TL D
Sbjct 816 QYSGGLVLEPKKGLYETLILLLDFNSLYPSIIQEYNICYTTLEYSKDSD--------EQL 867
Query 98 SSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANS 157
S P+ T ++ GVLP + +L+ R+ K+ M ++ K ++ RQ+ALK+TANS
Sbjct 868 SVPQSTDIE-----GVLPREIRKLVECRRDVKSLMKSERNEAKKKQMDIRQMALKLTANS 922
Query 158 VYG 160
+YG
Sbjct 923 MYG 925
> cpv:cgd3_4290 DNA polymerase alpha catalytic subunit ; K02320
DNA polymerase alpha subunit A [EC:2.7.7.7]
Length=1488
Score = 80.5 bits (197), Expect = 2e-15, Method: Composition-based stats.
Identities = 51/131 (38%), Positives = 70/131 (53%), Gaps = 12/131 (9%)
Query 35 RDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDV-----QTLQ 89
+++ Y G VLEP+ G Y+ I LDF SLYPSI+ NIC++T + TD+ +T +
Sbjct 835 KEETYSGGLVLEPITGLYDSFILLLDFNSLYPSIIQEFNICFTTR-EPTDLGPDATETFE 893
Query 90 QDTLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQL 149
+L SS VLP I+E L+ R+ K + KS L RQL
Sbjct 894 SSSLIDRNSSSNQ------FDSTVLPGILESLVKRRRHIKEILKNMPSGTRKSQLEIRQL 947
Query 150 ALKITANSVYG 160
ALK+TANS+YG
Sbjct 948 ALKLTANSLYG 958
> ath:AT5G67100 ICU2; ICU2 (INCURVATA2); DNA-directed DNA polymerase;
K02320 DNA polymerase alpha subunit A [EC:2.7.7.7]
Length=1492
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 48/122 (39%), Positives = 68/122 (55%), Gaps = 17/122 (13%)
Query 39 YEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTS 98
Y G VLEP +G Y+ + LDF SLYPSI+ +NIC++T+ + D
Sbjct 897 YAGGLVLEPKRGLYDKYVLLLDFNSLYPSIIQEYNICFTTIPRSED-------------G 943
Query 99 SPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSV 158
PR + +S G+LP ++E L++ RK K +M K E L+ RQ ALK+TANS+
Sbjct 944 VPR---LPSSQTPGILPKLMEHLVSIRKSVKLKMKK-ETGLKYWELDIRQQALKLTANSM 999
Query 159 YG 160
YG
Sbjct 1000 YG 1001
> bbo:BBOV_IV007060 23.m06311; DNA polymerase alpha subunit (EC:2.7.7.7);
K02320 DNA polymerase alpha subunit A [EC:2.7.7.7]
Length=1483
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 47/125 (37%), Positives = 67/125 (53%), Gaps = 21/125 (16%)
Query 38 KYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYST--LVKGTDVQTLQQDTLTH 95
YEG VLEP G Y+ + LDF SLYPSI+ N+C++T L++ VQ L + T
Sbjct 796 NYEGGLVLEPETGLYDNFVLLLDFNSLYPSIIQEFNVCFTTVKLLEDGSVQVLTETT--- 852
Query 96 TTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITA 155
G+LP I++ L+ R KA ++ ++ K+ L RQLALK+ A
Sbjct 853 ----------------GMLPQILKRLVELRASVKAAIAVEKNDSRKAQLGVRQLALKLVA 896
Query 156 NSVYG 160
NS+YG
Sbjct 897 NSLYG 901
> tpv:TP03_0202 DNA polymerase alpha (EC:2.7.7.7); K02320 DNA
polymerase alpha subunit A [EC:2.7.7.7]
Length=1451
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/127 (35%), Positives = 65/127 (51%), Gaps = 18/127 (14%)
Query 34 DRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTL 93
D YEG V EP+ G Y+ I LDF SLYPSI+ +NIC++T V + + D +
Sbjct 729 DDTKSYEGGLVFEPISGLYDNFILLLDFNSLYPSIIQEYNICFTTTVANGEESVVILDNV 788
Query 94 THTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKI 153
GVLP I++ L+ R K + +++ + L+ RQLALK+
Sbjct 789 ------------------GVLPSILKRLVELRLNIKNIIKGEKNETRRVQLSTRQLALKL 830
Query 154 TANSVYG 160
ANS+YG
Sbjct 831 IANSIYG 837
> sce:YNL102W POL1, CDC17, CRT5, HPR3; Catalytic subunit of the
DNA polymerase I alpha-primase complex, required for the initiation
of DNA replication during mitotic DNA synthesis and
premeiotic DNA synthesis (EC:2.7.7.7); K02320 DNA polymerase
alpha subunit A [EC:2.7.7.7]
Length=1468
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 65/123 (52%), Gaps = 12/123 (9%)
Query 38 KYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTT 97
KY+G V EP KG + + +DF SLYPSI+ NIC++T+ + ++ +
Sbjct 842 KYQGGLVFEPEKGLHKNYVLVMDFNSLYPSIIQEFNICFTTVDRN-------KEDIDELP 894
Query 98 SSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANS 157
S P + V +GVLP ++ L+ R+ K M D + + RQ ALK+TANS
Sbjct 895 SVP-----PSEVDQGVLPRLLANLVDRRREVKKVMKTETDPHKRVQCDIRQQALKLTANS 949
Query 158 VYG 160
+YG
Sbjct 950 MYG 952
> tgo:TGME49_017910 DNA polymerase alpha catalytic subunit (EC:2.7.7.7);
K02320 DNA polymerase alpha subunit A [EC:2.7.7.7]
Length=2017
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 45/148 (30%), Positives = 69/148 (46%), Gaps = 16/148 (10%)
Query 29 KRVGGDRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTL 88
++VG Y G VLEP G Y+ + LDF SLYPSI+ +N+C+ST+ + +
Sbjct 1097 EKVGARAKASYAGGLVLEPRVGLYDTFVLLLDFNSLYPSIIQEYNVCFSTVKRPVRTRIS 1156
Query 89 QQDTLTHTTSSPRHTFVKASVRR----------------GVLPMIVEELIAARKVAKAEM 132
+ F + R G+LP I+ +L+ R+ KA M
Sbjct 1157 RVQATNGPDGEIADAFREPDDRNLPHAAEEDVQGIASTPGLLPRILRQLVQRRRAVKASM 1216
Query 133 SKAEDKFMKSVLNGRQLALKITANSVYG 160
+ ++ +QLALK+TANS+YG
Sbjct 1217 KGESNPVKRATQEIKQLALKLTANSIYG 1244
> dre:317740 pola1, cb678, pola; polymerase (DNA directed), alpha
1; K02320 DNA polymerase alpha subunit A [EC:2.7.7.7]
Length=1470
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 69/125 (55%), Gaps = 8/125 (6%)
Query 39 YEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTS 98
Y G VL+P GFY+ I LDF SLYPSI+ NIC++T+ +G T ++ +
Sbjct 848 YAGGLVLDPKVGFYDKFILLLDFNSLYPSIIQEFNICFTTVERGA-TNTRKKTEVGDEDE 906
Query 99 SPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAE---DKFMKSVLNGRQLALKITA 155
P S+ G+LP + +L+ R+ K M + + D +++ + RQ ALK+TA
Sbjct 907 IPE--LPDQSLEMGILPKEIRKLVERRRQVKQLMKEPDLNPDLYLQYDI--RQKALKLTA 962
Query 156 NSVYG 160
NS+YG
Sbjct 963 NSMYG 967
> mmu:18968 Pola1, AW321876, Pola; polymerase (DNA directed),
alpha 1 (EC:2.7.7.7); K02320 DNA polymerase alpha subunit A
[EC:2.7.7.7]
Length=1465
Score = 72.0 bits (175), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 49/131 (37%), Positives = 71/131 (54%), Gaps = 8/131 (6%)
Query 33 GDRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGT-DVQTLQQD 91
G + Y G VL+P GFY+ I LDF SLYPSI+ NIC++T+ + T +VQ +D
Sbjct 837 GRKKATYAGGLVLDPKVGFYDKFILLLDFNSLYPSIIQEFNICFTTVQRVTSEVQKATED 896
Query 92 TLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVL--NGRQL 149
++ G+LP + +L+ RK K ++ K +D VL + RQ
Sbjct 897 EEQEQIPE----LPDPNLEMGILPREIRKLVERRKQVK-QLMKQQDLNPDLVLQYDIRQK 951
Query 150 ALKITANSVYG 160
ALK+TANS+YG
Sbjct 952 ALKLTANSMYG 962
> hsa:5422 POLA1, DKFZp686K1672, POLA, p180; polymerase (DNA directed),
alpha 1, catalytic subunit (EC:2.7.7.7); K02320 DNA
polymerase alpha subunit A [EC:2.7.7.7]
Length=1462
Score = 72.0 bits (175), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 47/131 (35%), Positives = 71/131 (54%), Gaps = 8/131 (6%)
Query 33 GDRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVK-GTDVQTLQQD 91
G + Y G VL+P GFY+ I LDF SLYPSI+ NIC++T+ + ++ Q + +D
Sbjct 833 GRKKAAYAGGLVLDPKVGFYDKFILLLDFNSLYPSIIQEFNICFTTVQRVASEAQKVTED 892
Query 92 TLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVL--NGRQL 149
S+ G+LP + +L+ RK K ++ K +D +L + RQ
Sbjct 893 GEQEQIPE----LPDPSLEMGILPREIRKLVERRKQVK-QLMKQQDLNPDLILQYDIRQK 947
Query 150 ALKITANSVYG 160
ALK+TANS+YG
Sbjct 948 ALKLTANSMYG 958
> xla:398200 pola1, pola; polymerase (DNA directed), alpha 1,
catalytic subunit (EC:2.7.7.7); K02320 DNA polymerase alpha
subunit A [EC:2.7.7.7]
Length=1458
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 48/132 (36%), Positives = 69/132 (52%), Gaps = 25/132 (18%)
Query 39 YEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTL-------VKGTDVQTLQQD 91
Y G VLEP GFY+ I LDF SLYPSI+ +NIC++T+ KG D QD
Sbjct 838 YAGGLVLEPKVGFYDKFILLLDFNSLYPSIIQEYNICFTTVHREAPSTQKGED-----QD 892
Query 92 TLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAE---DKFMKSVLNGRQ 148
+ S + G+LP + +L+ R+ K M + + D +++ + RQ
Sbjct 893 EIPELPHS--------DLEMGILPREIRKLVERRRHVKQLMKQPDLNPDLYLQYDI--RQ 942
Query 149 LALKITANSVYG 160
ALK+TANS+YG
Sbjct 943 KALKLTANSMYG 954
> tgo:TGME49_064670 DNA polymerase zeta catalytic subunit, putative
(EC:2.7.7.7); K02350 DNA polymerase zeta subunit [EC:2.7.7.7]
Length=3952
Score = 69.3 bits (168), Expect = 6e-12, Method: Composition-based stats.
Identities = 41/95 (43%), Positives = 56/95 (58%), Gaps = 3/95 (3%)
Query 2 LTRGQQIKVTSQLLRKCRQLHYLV--PSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTL 59
LTRG Q KV S +++ ++L YL PS +V ++ VLEP GFY P+ L
Sbjct 2921 LTRGSQFKVESVMIQATKRLDYLFVSPSQTQVT-EQPAPLCIPLVLEPKSGFYWSPVLVL 2979
Query 60 DFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLT 94
DF SLYPSI++A+NICYST + ++ Q T T
Sbjct 2980 DFRSLYPSIVIANNICYSTALGSVEIDPTVQATKT 3014
Score = 42.7 bits (99), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 5/62 (8%)
Query 104 FVKASVRRGVLPMIVEELIAAR----KVAKAEMSKAEDKFMKSVLNGRQLALKITANSVY 159
FV VR+G+ P ++ E++ R K K K E+ ++ +LN RQ LK+ AN Y
Sbjct 3099 FVSRRVRKGIFPQVLSEILQTRIMVQKAIKKYKGKVEEGLLR-LLNYRQYGLKMIANVSY 3157
Query 160 GY 161
GY
Sbjct 3158 GY 3159
> bbo:BBOV_III010120 17.m07879; DNA polymerase family B family
protein; K02350 DNA polymerase zeta subunit [EC:2.7.7.7]
Length=1478
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 52/183 (28%), Positives = 84/183 (45%), Gaps = 28/183 (15%)
Query 2 LTRGQQIKVTSQLLRKCRQLHYLVPS-VKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
+ RG Q V S LLR + YL+PS +R ++ V++P G + P++ LD
Sbjct 847 IVRGSQYHVESVLLRFTKSYGYLLPSPTQRQVHEQRPSVAIPIVMQPQSGLHLAPVAILD 906
Query 61 FASLYPSIMMAHNICYST------------------LVKGTDVQTLQQDTLTHTTS---- 98
F SLY I +A+NICYST ++ + + +D H T
Sbjct 907 FQSLYSCITIAYNICYSTCLGLLSEHKKNSGSIKLGIMTYYPEEGILEDITEHRTDKGIG 966
Query 99 ---SPRHT-FVKASVRRGVLPMIVEELIAARKVAKAEMSKAE-DKFMKSVLNGRQLALKI 153
+P FV VR G+LPM+++ ++ +R+ K M + + D L+ Q LK+
Sbjct 967 VHITPNGVMFVDKRVREGILPMMLKVVLQSRRKIKEAMKQPDMDAISLHKLDREQHGLKM 1026
Query 154 TAN 156
+N
Sbjct 1027 LSN 1029
> pfa:PFD0590c DNA polymerase alpha; K02320 DNA polymerase alpha
subunit A [EC:2.7.7.7]
Length=1912
Score = 55.5 bits (132), Expect = 7e-08, Method: Composition-based stats.
Identities = 26/49 (53%), Positives = 36/49 (73%), Gaps = 1/49 (2%)
Query 38 KYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTL-VKGTDV 85
KY G VL+PL G+Y+ + LDF SLYPSI++ +N+C+STL +K DV
Sbjct 1082 KYLGGLVLDPLCGYYDTFVLYLDFNSLYPSIIIEYNVCFSTLKLKNCDV 1130
Score = 39.3 bits (90), Expect = 0.006, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 32/49 (65%), Gaps = 0/49 (0%)
Query 112 GVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYG 160
G+LP I++ L+ R V K +S ++K K +L + L++K+ +NS+YG
Sbjct 1207 GILPCILKSLVEKRSVIKKLISNEKNKEKKELLLIQSLSIKLISNSIYG 1255
> pfa:PF10_0362 DNA polymerase zeta catalytic subunit, putative;
K02350 DNA polymerase zeta subunit [EC:2.7.7.7]
Length=2240
Score = 52.0 bits (123), Expect = 8e-07, Method: Composition-based stats.
Identities = 30/80 (37%), Positives = 44/80 (55%), Gaps = 3/80 (3%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLV--PSVKRVGGDRDDKYEGATVLEPLKGFYNMPIST 58
L+ RG Q + S LL+ + +Y++ PS K + D+ +L+P P+
Sbjct 1046 LINRGSQYIIESFLLKMSIKYNYVLYSPSNKEIF-DQRPILHTPLILQPKSSINFFPLLV 1104
Query 59 LDFASLYPSIMMAHNICYST 78
DF SLYPSI++A NICYST
Sbjct 1105 FDFQSLYPSILIAFNICYST 1124
Score = 35.4 bits (80), Expect = 0.084, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 1/64 (1%)
Query 98 SSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANS 157
+S +VK R+G+ P+ +E+++ R + K M E++ K LN R LK+ N
Sbjct 1328 TSNNTIYVKKHKRKGICPLFLEDILKTRIMLKRCMGMYEERVNKK-LNERMGKLKLILNV 1386
Query 158 VYGY 161
GY
Sbjct 1387 ATGY 1390
> eco:b0060 polB, dinA, ECK0061, JW0059; DNA polymerase II (EC:2.7.7.7);
K02336 DNA polymerase II [EC:2.7.7.7]
Length=783
Score = 36.6 bits (83), Expect = 0.039, Method: Composition-based stats.
Identities = 35/117 (29%), Positives = 53/117 (45%), Gaps = 12/117 (10%)
Query 41 GATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSP 100
G V++ G Y+ + LD+ SLYPSI+ I LV+G + Q H+T
Sbjct 401 GGYVMDSRPGLYD-SVLVLDYKSLYPSIIRTFLIDPVGLVEG-----MAQPDPEHSTEG- 453
Query 101 RHTFVKA--SVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITA 155
F+ A S + LP IV + R AK + +K + +K ++N L TA
Sbjct 454 ---FLDAWFSREKHCLPEIVTNIWHGRDEAKRQGNKPLSQALKIIMNAFYGVLGTTA 507
> tgo:TGME49_039090 surface antigen, putative
Length=294
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 50/134 (37%), Gaps = 26/134 (19%)
Query 8 IKVTSQLLR-KCRQLHYLVPSVKRVGGDRDDKYEGATVLEPL--------------KGFY 52
+KVT L KC+ L P V+ V D D K L L G Y
Sbjct 168 VKVTGNALSLKCKDLTLDPPDVQNVYDDEDGKCASKVALTSLVDGSLAAVAAEQEDNGEY 227
Query 53 NMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDT---LTHTTSSPRHTFVKASV 109
+ IS L P+ A ++CY + K + Q T L T +S F AS+
Sbjct 228 ALSISEL------PA--DAKHLCYKCVAKQSRTQAGSASTECMLKLTVTSGAAAFATASI 279
Query 110 RRGVLPMIVEELIA 123
G+L + L A
Sbjct 280 SVGILASLASFLSA 293
> sce:YMR162C DNF3; Aminophospholipid translocase (flippase) that
maintains membrane lipid asymmetry in post-Golgi secretory
vesicles; localizes to the trans-Golgi network; likely involved
in protein transport; type 4 P-type ATPase (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1656
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 43/88 (48%), Gaps = 13/88 (14%)
Query 26 PSVKRV-GGDRDDKYEGATVLEPLKGFYNMPISTLD---FASLYPSIMMAHN-------- 73
PS++ + G D + A+V+ P + F S+ D F YP+ + +
Sbjct 676 PSMRSLFGKDNSHLSKQASVISPSETFSENIKSSFDLIQFIQRYPTALFSQKAKFFFLSL 735
Query 74 -ICYSTLVKGTDVQTLQQDTLTHTTSSP 100
+C+S L K T +++ +D++ + +SSP
Sbjct 736 ALCHSCLPKKTHNESIGEDSIEYQSSSP 763
> ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=567
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 32/71 (45%), Gaps = 13/71 (18%)
Query 9 KVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSI 68
K LR CRQLH S + GD TVL P+KG +P T +SL+P
Sbjct 11 KTILSFLRPCRQLH----STPKSTGD-------VTVLSPVKGRRRLP--TCWSSSLFPLA 57
Query 69 MMAHNICYSTL 79
+ A ++ L
Sbjct 58 IAASATSFAYL 68
> ath:AT1G49180 protein kinase family protein
Length=408
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/20 (65%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 1 LLTRGQQIKVTSQLLRKCRQ 20
L++RG+ IKV SQLLRK +Q
Sbjct 334 LISRGESIKVLSQLLRKAKQ 353
> ath:AT5G27470 seryl-tRNA synthetase / serine--tRNA ligase (EC:6.1.1.11);
K01875 seryl-tRNA synthetase [EC:6.1.1.11]
Length=451
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 14 LLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFY 52
++ KC QL + +V G+ DDKY AT +PL ++
Sbjct 209 VMAKCAQLAQFDEELYKVTGEGDDKYLIATAEQPLCAYH 247
> hsa:55214 LEPREL1, FLJ10718, MLAT4, P3H2; leprecan-like 1 (EC:1.14.11.7)
Length=527
Score = 28.9 bits (63), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 9/46 (19%)
Query 16 RKCRQLHYLVPSVKRVG-GDR--------DDKYEGATVLEPLKGFY 52
+CR+LH + + VG G R ++K+EGATVL+ LK Y
Sbjct 293 EQCRELHSVASGIMLVGDGYRGKTSPHTPNEKFEGATVLKALKSGY 338
> mmu:210530 Leprel1, 4832416N06, AW553532, Mlat4, P3H2; leprecan-like
1 (EC:1.14.11.7)
Length=703
Score = 28.9 bits (63), Expect = 7.8, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 9/45 (20%)
Query 17 KCRQLHYLVPSVKRVG-GDR--------DDKYEGATVLEPLKGFY 52
+CR+LH + + VG G R ++K+EGATVL+ LK Y
Sbjct 470 QCRELHSVANGIMLVGDGYRGKTSPHTPNEKFEGATVLKALKFGY 514
> hsa:4297 MLL, ALL-1, CXXC7, FLJ11783, HRX, HTRX1, KMT2A, MLL/GAS7,
MLL1A, TET1-MLL, TRX1; myeloid/lymphoid or mixed-lineage
leukemia (trithorax homolog, Drosophila) (EC:2.1.1.43);
K09186 histone-lysine N-methyltransferase MLL1 [EC:2.1.1.43]
Length=3972
Score = 28.9 bits (63), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 37/88 (42%), Gaps = 13/88 (14%)
Query 49 KGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKAS 108
KG N+ + LD PS+ +C S T T+ H+TSS +A
Sbjct 964 KGRGNLEKTNLDLGPTAPSLEKEKTLCLS---------TPSSSTVKHSTSSIGSMLAQAD 1014
Query 109 VRRGVLPMIVEELIAARKVAKAEMSKAE 136
LPM + + + K AKA++ K E
Sbjct 1015 ----KLPMTDKRVASLLKKAKAQLCKIE 1038
Lambda K H
0.320 0.133 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3702936828
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40