bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1180_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
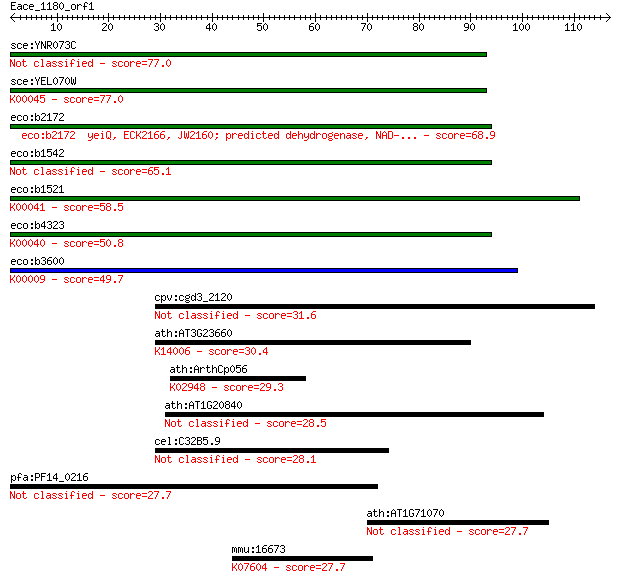
sce:YNR073C Putative mannitol dehydrogenase (EC:1.1.-.-) 77.0 1e-14
sce:YEL070W DSF1; Deletion suppressor of mpt5 mutation (EC:1.1... 77.0 1e-14
eco:b2172 yeiQ, ECK2166, JW2160; predicted dehydrogenase, NAD-... 68.9 3e-12
eco:b1542 ydfI, ECK1535, JW1535; predicted mannonate dehydroge... 65.1 6e-11
eco:b1521 uxaB, ECK1514, JW1514; altronate oxidoreductase, NAD... 58.5 5e-09
eco:b4323 uxuB, ECK4314, JW4286; D-mannonate oxidoreductase, N... 50.8 9e-07
eco:b3600 mtlD, ECK3589, JW3574; mannitol-1-phosphate dehydrog... 49.7 2e-06
cpv:cgd3_2120 myosin fused to 3 IQ motifs (that interact with ... 31.6 0.69
ath:AT3G23660 transport protein, putative; K14006 protein tran... 30.4 1.4
ath:ArthCp056 rps11; 30S ribosomal protein S11; K02948 small s... 29.3 3.2
ath:AT1G20840 TMT1; TMT1 (TONOPLAST MONOSACCHARIDE TRANSPORTER... 28.5 5.4
cel:C32B5.9 fbxc-7; F-box C protein family member (fbxc-7) 28.1 6.6
pfa:PF14_0216 conserved Plasmodium protein, unknown function 27.7 8.5
ath:AT1G71070 glycosyltransferase family 14 protein / core-2/I... 27.7 9.2
mmu:16673 Krt36, HRa-1, K36, Krt1-22, Krt1-5, MHRa-1; keratin ... 27.7 9.9
> sce:YNR073C Putative mannitol dehydrogenase (EC:1.1.-.-)
Length=502
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 38/94 (40%), Positives = 57/94 (60%), Gaps = 2/94 (2%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRDVRVSGFLRSFM-DEVTPCLEDLK 59
V SYE M+L+LLNGGHS + Y+ L GY ++ E + D ++ ++R M +EV P L +
Sbjct 297 VDSYELMKLRLLNGGHSAMGYLGYLAGYTYIHEVVNDPTINKYIRVLMREEVIPLLPKVP 356
Query 60 -VDVERYKERIVERFGNAFVKDELQRVAQDGSSK 92
VD E Y ++ERF N ++D + R+ GS K
Sbjct 357 GVDFEEYTASVLERFSNPAIQDTVARICLMGSGK 390
> sce:YEL070W DSF1; Deletion suppressor of mpt5 mutation (EC:1.1.-.-);
K00045 mannitol 2-dehydrogenase [EC:1.1.1.67]
Length=502
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 38/94 (40%), Positives = 57/94 (60%), Gaps = 2/94 (2%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRDVRVSGFLRSFM-DEVTPCLEDLK 59
V SYE M+L+LLNGGHS + Y+ L GY ++ E + D ++ ++R M +EV P L +
Sbjct 297 VDSYELMKLRLLNGGHSAMGYLGYLAGYTYIHEVVNDPTINKYIRVLMREEVIPLLPKVP 356
Query 60 -VDVERYKERIVERFGNAFVKDELQRVAQDGSSK 92
VD E Y ++ERF N ++D + R+ GS K
Sbjct 357 GVDFEEYTASVLERFSNPAIQDTVARICLMGSGK 390
> eco:b2172 yeiQ, ECK2166, JW2160; predicted dehydrogenase, NAD-dependent;
K00540 [EC:1.-.-.-]
Length=488
Score = 68.9 bits (167), Expect = 3e-12, Method: Composition-based stats.
Identities = 33/94 (35%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRDVRVSGFLRSFM-DEVTPCLEDLK 59
V +EEM+L++LNG HS +AY+ L G+ + + ++D R+ M DE P L+
Sbjct 284 VLPWEEMKLRMLNGSHSFLAYLGYLSGFAHISDCMQDRAFRHAARTLMLDEQAPTLQIKD 343
Query 60 VDVERYKERIVERFGNAFVKDELQRVAQDGSSKF 93
VD+ +Y ++++ RF N +K + ++A DGS K
Sbjct 344 VDLTQYADKLIARFANPALKHKTWQIAMDGSQKL 377
> eco:b1542 ydfI, ECK1535, JW1535; predicted mannonate dehydrogenase
Length=486
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 55/95 (57%), Gaps = 3/95 (3%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRD--VRVSGFLRSFMDEVTPCLEDL 58
V YEEM+L++LNG HS +AY+ L GY+ +++ + D R + + + E P L+
Sbjct 281 VLPYEEMKLRMLNGSHSFLAYLGYLAGYQHINDCMEDEHYRYAAY-GLMLQEQAPTLKVQ 339
Query 59 KVDVERYKERIVERFGNAFVKDELQRVAQDGSSKF 93
VD++ Y R++ R+ N ++ ++A DGS K
Sbjct 340 GVDLQDYANRLIARYSNPALRHRTWQIAMDGSQKL 374
> eco:b1521 uxaB, ECK1514, JW1514; altronate oxidoreductase, NAD-dependent
(EC:1.1.1.58); K00041 tagaturonate reductase [EC:1.1.1.58]
Length=483
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 33/111 (29%), Positives = 61/111 (54%), Gaps = 3/111 (2%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRDVRVSGFL-RSFMDEVTPCLEDLK 59
++ Y+E ++ +LNG H+ + VA G V EA+ D + F+ ++ +E+ P L+ +
Sbjct 274 IKPYKERKVAILNGAHTALVPVAFQAGLDTVGEAMNDAEICAFVEKAIYEEIIPVLDLPR 333
Query 60 VDVERYKERIVERFGNAFVKDELQRVAQDGSSKFYSTTRNSTLLLLERKKN 110
++E + + RF N ++K +L +A +G +KF TR LL +K N
Sbjct 334 DELESFASAVTGRFRNPYIKHQLLSIALNGMTKF--RTRILPQLLAGQKAN 382
> eco:b4323 uxuB, ECK4314, JW4286; D-mannonate oxidoreductase,
NAD-binding (EC:1.1.1.57); K00040 fructuronate reductase [EC:1.1.1.57]
Length=486
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 50/96 (52%), Gaps = 4/96 (4%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRD--VRVSGFLRSFMDEVTPCLEDL 58
V +E M+L++LNG HS +AY+ L GY + + + + R + F M E P L
Sbjct 284 VVPFEMMKLRMLNGSHSFLAYLGYLGGYETIADTVTNPAYRKAAFAL-MMQEQAPTLSMP 342
Query 59 K-VDVERYKERIVERFGNAFVKDELQRVAQDGSSKF 93
+ D+ Y ++ERF N ++ ++A DGS K
Sbjct 343 EGTDLNAYATLLIERFSNPSLRHRTWQIAMDGSQKL 378
> eco:b3600 mtlD, ECK3589, JW3574; mannitol-1-phosphate dehydrogenase,
NAD(P)-binding (EC:1.1.1.17); K00009 mannitol-1-phosphate
5-dehydrogenase [EC:1.1.1.17]
Length=382
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/102 (26%), Positives = 56/102 (54%), Gaps = 4/102 (3%)
Query 1 VQSYEEMRLKLLNGGHSCVAYVATLLGYRFVDEALRDVRVSGFLRSFMDEVTPCL-EDLK 59
+ ++ E +L LN GH+ AY+ L G++ + +A+ D ++ ++ M+E L +
Sbjct 205 LMAFVERKLFTLNTGHAITAYLGKLAGHQTIRDAILDEKIRAVVKGAMEESGAVLIKRYG 264
Query 60 VDVER---YKERIVERFGNAFVKDELQRVAQDGSSKFYSTTR 98
D ++ Y ++I+ RF N ++KD+++RV + K + R
Sbjct 265 FDADKHAAYIQKILGRFENPYLKDDVERVGRQPLRKLSAGDR 306
> cpv:cgd3_2120 myosin fused to 3 IQ motifs (that interact with
calmodulin) and Rcc1 domain
Length=2083
Score = 31.6 bits (70), Expect = 0.69, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 8/89 (8%)
Query 29 RFVDEALRDVRVSGFLRSFMDEVTPCLEDLKVDVERYKERIVERFGNAFVKDELQRVAQD 88
R V+E + +++ S + + TPC E ++D++ K I +F +F +EL + +
Sbjct 556 RRVEEFISEIQKDPTESSLLHKPTPCDEFGRIDLDPSKS-IYSKFVRSFTGNELLKANDE 614
Query 89 ----GSSKFYSTTRNSTLLLLERKKNCLK 113
G SKF N LL+RKK K
Sbjct 615 FITKGESKFQEEPYNE---LLKRKKQICK 640
> ath:AT3G23660 transport protein, putative; K14006 protein transport
protein SEC23
Length=765
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 8/67 (11%)
Query 29 RFVDEALRDVRVSG----FLRSFMDEVTPCLEDLKVDVERYKERIV--ERFGNAFVKDEL 82
+F D + + G S +D+V + ++KV VER +V E FG++ KD
Sbjct 332 KFYDSIAKQLVTQGHVLDLFASALDQVG--VAEMKVAVERTGGLVVLSESFGHSVFKDSF 389
Query 83 QRVAQDG 89
+RV +DG
Sbjct 390 KRVFEDG 396
> ath:ArthCp056 rps11; 30S ribosomal protein S11; K02948 small
subunit ribosomal protein S11
Length=138
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 32 DEALRDVRVSGFLRSFMDEVTPCLED 57
D ALR +R SG L SF+ +VTP +
Sbjct 103 DAALRAIRRSGILLSFVRDVTPMPHN 128
> ath:AT1G20840 TMT1; TMT1 (TONOPLAST MONOSACCHARIDE TRANSPORTER1);
carbohydrate transmembrane transporter/ nucleoside transmembrane
transporter/ sugar:hydrogen symporter
Length=734
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 0/73 (0%)
Query 31 VDEALRDVRVSGFLRSFMDEVTPCLEDLKVDVERYKERIVERFGNAFVKDELQRVAQDGS 90
+DEA R ++ DE+ +E L + E+ E ++ + D L+ V +DG
Sbjct 196 MDEAKRVLQQLCGREDVTDEMALLVEGLDIGGEKTMEDLLVTLEDHEGDDTLETVDEDGQ 255
Query 91 SKFYSTTRNSTLL 103
+ Y T N + L
Sbjct 256 MRLYGTHENQSYL 268
> cel:C32B5.9 fbxc-7; F-box C protein family member (fbxc-7)
Length=470
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 1/46 (2%)
Query 29 RFVDEALRDVRVSGFLRSF-MDEVTPCLEDLKVDVERYKERIVERF 73
R +D + R +GF S M+E E LK + +KERI+ERF
Sbjct 128 RLIDNWIEVGREAGFYYSLEMNEEIRAREILKTVMINHKERIIERF 173
> pfa:PF14_0216 conserved Plasmodium protein, unknown function
Length=641
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 20/77 (25%), Positives = 38/77 (49%), Gaps = 6/77 (7%)
Query 1 VQSYEEMRL--KLLNGGHSCVAYVATLLGYRFV----DEALRDVRVSGFLRSFMDEVTPC 54
+ YE +RL N +SC++ + Y F+ + L + R+S F F+D +
Sbjct 490 MNDYEMLRLICSYSNDKNSCISKKMSYFVYLFLKNLPQKELMNFRISDFASPFLDFINAK 549
Query 55 LEDLKVDVERYKERIVE 71
LED K +++ + ++E
Sbjct 550 LEDTKKYIKQALQILLE 566
> ath:AT1G71070 glycosyltransferase family 14 protein / core-2/I-branching
enzyme family protein
Length=395
Score = 27.7 bits (60), Expect = 9.2, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 70 VERFGNAFVKDELQRVAQDGSSKFYSTTRNSTLLL 104
V FGN V ++ R++++G+SK ST ++LL
Sbjct 103 VNAFGNVDVLGKVDRLSENGASKIASTLHAVSILL 137
> mmu:16673 Krt36, HRa-1, K36, Krt1-22, Krt1-5, MHRa-1; keratin
36; K07604 type I keratin, acidic
Length=473
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 44 LRSFMDEVTPCLEDLKVDVERYKERIV 70
LR +DE+T C DL+ VE KE ++
Sbjct 199 LRRILDELTLCKADLEAQVESLKEELL 225
Lambda K H
0.322 0.138 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40