bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1182_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
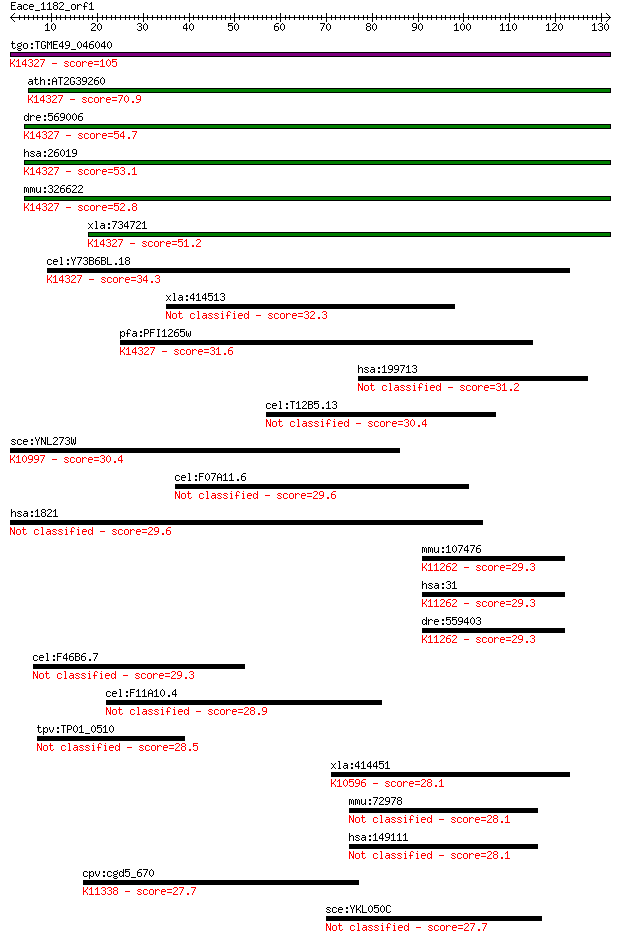
tgo:TGME49_046040 MIF4G domain-containing protein ; K14327 reg... 105 4e-23
ath:AT2G39260 RNA binding / binding / protein binding; K14327 ... 70.9 9e-13
dre:569006 upf2; UPF2 regulator of nonsense transcripts homolo... 54.7 8e-08
hsa:26019 UPF2, DKFZp434D222, HUPF2, KIAA1408, MGC138834, MGC1... 53.1 2e-07
mmu:326622 Upf2; UPF2 regulator of nonsense transcripts homolo... 52.8 3e-07
xla:734721 upf2, hupf2, rent2, smg-3; UPF2 regulator of nonsen... 51.2 9e-07
cel:Y73B6BL.18 smg-3; Suppressor with Morphological effect on ... 34.3 0.11
xla:414513 inf2, MGC81508; inverted formin, FH2 and WH2 domain... 32.3 0.42
pfa:PFI1265w conserved Plasmodium protein, unknown function; K... 31.6 0.64
hsa:199713 NLRP7, CLR19.4, FLJ94610, HYDM, MGC126470, MGC12647... 31.2 0.82
cel:T12B5.13 fbxa-23; F-box A protein family member (fbxa-23) 30.4 1.4
sce:YNL273W TOF1; Subunit of a replication-pausing checkpoint ... 30.4 1.5
cel:F07A11.6 din-1; DAF-12 Interacting Protein family member (... 29.6 2.4
hsa:1821 DRP2, MGC133255; dystrophin related protein 2 29.6
mmu:107476 Acaca, A530025K05Rik, Acac, Acc1, Gm738; acetyl-Coe... 29.3 3.2
hsa:31 ACACA, ACAC, ACC, ACC1, ACCA; acetyl-CoA carboxylase al... 29.3 3.2
dre:559403 acaca, im:7138837, si:ch211-199d18.1, wu:fj43d01; a... 29.3 3.4
cel:F46B6.7 ztf-7; Zinc finger Transcription Factor family mem... 29.3 3.6
cel:F11A10.4 mon-2; yeast MON (monensin-resistant) homolog fam... 28.9 4.1
tpv:TP01_0510 hypothetical protein 28.5 5.8
xla:414451 ube4a, MGC114906; ubiquitination factor E4A (UFD2 h... 28.1 7.1
mmu:72978 Cnih3, 2900075G08Rik; cornichon homolog 3 (Drosophila) 28.1 7.7
hsa:149111 CNIH3, CNIH-3, FLJ38993; cornichon homolog 3 (Droso... 28.1 7.7
cpv:cgd5_670 reptin52; reptin like TIP49 family AAA+ ATpase ; ... 27.7 8.7
sce:YKL050C Protein of unknown function; the YKL050W protein i... 27.7 9.3
> tgo:TGME49_046040 MIF4G domain-containing protein ; K14327 regulator
of nonsense transcripts 2
Length=1439
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 53/131 (40%), Positives = 81/131 (61%), Gaps = 0/131 (0%)
Query 1 AMAALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARL 60
M +L R+SQA+ A +DQIA D F ERLN K NR+ L I V+ T +Q +P ARL
Sbjct 657 GMDLMLLRLSQASEAHAVDQIAMDIFYERLNTKVNRAALTKAITSVSRTQLQLLPFYARL 716
Query 61 LAILRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLL 120
LAIL+ +K+ L+ LQ + +++ ++ PT++E +++ +RF+ E K LLP G +L
Sbjct 717 LAILKPFMKDVCGGVLDYLQGEMKRILEEKHPTELESKIKNVRFIAECTKFGLLPVGFVL 776
Query 121 NCCSSWLEDFT 131
+S +ED T
Sbjct 777 GAFNSLMEDMT 787
> ath:AT2G39260 RNA binding / binding / protein binding; K14327
regulator of nonsense transcripts 2
Length=1181
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 36/127 (28%), Positives = 73/127 (57%), Gaps = 2/127 (1%)
Query 5 LLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAIL 64
LL R+ + IDQ+ ++ LN+K NR +L+ + V T+++ + +R++A L
Sbjct 470 LLQRLPGCVSRDLIDQLTVEYCY--LNSKTNRKKLVKALFNVPRTSLELLAYYSRMVATL 527
Query 65 RKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCS 124
+K+ + +++L+ ++ L D +E ++R +RF+ EL K K++P GL+ +C
Sbjct 528 ASCMKDIPSMLVQMLEDEFNSLVHKKDQMNIETKIRNIRFIGELCKFKIVPAGLVFSCLK 587
Query 125 SWLEDFT 131
+ L++FT
Sbjct 588 ACLDEFT 594
> dre:569006 upf2; UPF2 regulator of nonsense transcripts homolog
(yeast); K14327 regulator of nonsense transcripts 2
Length=594
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 33/128 (25%), Positives = 58/128 (45%), Gaps = 1/128 (0%)
Query 4 ALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAI 63
A + ++ ID+ A DF + +N K NR +L+ + V + +P ARL+A
Sbjct 154 AFIQQLPNCINRDLIDKAAMDFCMN-MNTKSNRRKLVRALFTVPRQRLDLLPFYARLVAT 212
Query 64 LRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCC 123
L + + +L+ +R D +E + + +RF+ EL K K+ L+C
Sbjct 213 LHPCMSDVAEDLCSMLKGDFRFHIRKKDQINIETKNKTVRFIGELAKFKMFSKTDTLHCL 272
Query 124 SSWLEDFT 131
L DF+
Sbjct 273 KMLLSDFS 280
> hsa:26019 UPF2, DKFZp434D222, HUPF2, KIAA1408, MGC138834, MGC138835,
RENT2, smg-3; UPF2 regulator of nonsense transcripts
homolog (yeast); K14327 regulator of nonsense transcripts
2
Length=1272
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 58/128 (45%), Gaps = 1/128 (0%)
Query 4 ALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAI 63
A L ++ ID+ A DF + +N K NR +L+ + V + +P ARL+A
Sbjct 570 AFLQQLPNCVNRDLIDKAAMDFCMN-MNTKANRKKLVRALFIVPRQRLDLLPFYARLVAT 628
Query 64 LRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCC 123
L + + +L+ +R D +E + + +RF+ EL K K+ L+C
Sbjct 629 LHPCMSDVAEDLCSMLRGDFRFHVRKKDQINIETKNKTVRFIGELTKFKMFTKNDTLHCL 688
Query 124 SSWLEDFT 131
L DF+
Sbjct 689 KMLLSDFS 696
> mmu:326622 Upf2; UPF2 regulator of nonsense transcripts homolog
(yeast); K14327 regulator of nonsense transcripts 2
Length=1269
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 58/128 (45%), Gaps = 1/128 (0%)
Query 4 ALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAI 63
A L ++ ID+ A DF + +N K NR +L+ + V + +P ARL+A
Sbjct 568 AFLQQLPNCVNRDLIDKAAMDFCM-NMNTKANRKKLVRALFIVPRQRLDLLPFYARLVAT 626
Query 64 LRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCC 123
L + + +L+ +R D +E + + +RF+ EL K K+ L+C
Sbjct 627 LHPCMSDVAEDLCSMLRGDFRFHVRKKDQINIETKNKTVRFIGELTKFKMFTKNDTLHCL 686
Query 124 SSWLEDFT 131
L DF+
Sbjct 687 KMLLSDFS 694
> xla:734721 upf2, hupf2, rent2, smg-3; UPF2 regulator of nonsense
transcripts homolog; K14327 regulator of nonsense transcripts
2
Length=1264
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 54/114 (47%), Gaps = 1/114 (0%)
Query 18 IDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKEETNQALE 77
ID+ A DF + +N K NR +L+ + V + +P ARL+A L + +
Sbjct 576 IDKAAMDFCM-NMNTKANRRKLVRALFIVPRQRLDLLPFYARLVATLHPCMSDIAEDLCS 634
Query 78 LLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCSSWLEDFT 131
+L+ +R D +E + + +RF+ EL K K+ L+C L DF+
Sbjct 635 MLKGDFRFHVRKKDQINIETKNKTVRFIGELAKFKMFNKTDTLHCLKMLLSDFS 688
> cel:Y73B6BL.18 smg-3; Suppressor with Morphological effect on
Genitalia family member (smg-3); K14327 regulator of nonsense
transcripts 2
Length=1142
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 56/119 (47%), Gaps = 10/119 (8%)
Query 9 ISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLL 68
+S+ +T Q AA +F+ LN K R L+ ++ + + +P ARL+A L ++
Sbjct 446 VSKYSTDQ-----AAIYFVSNLNNKGCRKRLVKLMIDPPPSRIDVVPFYARLVATLENVM 500
Query 69 KEETNQALELLQSKWRQLADDCDPT-----KVEERVRVLRFVCELIKLKLLPPGLLLNC 122
+ T + + L K+R + KVE ++ + + EL+K ++ L+C
Sbjct 501 PDLTTEIVTQLLEKFRGFLQQKPSSATAIIKVESKMVCVMMIAELMKFGVISRAEGLSC 559
> xla:414513 inf2, MGC81508; inverted formin, FH2 and WH2 domain
containing
Length=1099
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 36/71 (50%), Gaps = 11/71 (15%)
Query 35 NRSELIMHILFVAETNVQCMPCCARLLAILRKLLKEETN--------QALELLQSKWRQL 86
N + + LF V C P +LL+IL+ LL+ + + +ALE+L ++ L
Sbjct 269 NNHQEVFSTLF---NKVSCSPLSVQLLSILQGLLQLDQSHPTSPLLWEALEVLVNRAVLL 325
Query 87 ADDCDPTKVEE 97
ADDC VEE
Sbjct 326 ADDCQNNNVEE 336
> pfa:PFI1265w conserved Plasmodium protein, unknown function;
K14327 regulator of nonsense transcripts 2
Length=1754
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 21/90 (23%), Positives = 46/90 (51%), Gaps = 1/90 (1%)
Query 25 FFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKEETNQALELLQSKWR 84
FL N K+ R + +I+++ + + +P R + I+ K K+ ++ L+
Sbjct 1005 LFLLNYNTKKKRKIIANNIIYINKVTLNLIPYYCRYICIINKYAKDMYPIIIDELKKITE 1064
Query 85 QLADDCDPTKVEERVRVLRFVCELIKLKLL 114
++ + P + ++ + +++VCEL K KLL
Sbjct 1065 KIIKEKVPCE-NKKTKCIKYVCELTKFKLL 1093
> hsa:199713 NLRP7, CLR19.4, FLJ94610, HYDM, MGC126470, MGC126471,
NALP7, NOD12, PAN7, PYPAF3; NLR family, pyrin domain containing
7
Length=1037
Score = 31.2 bits (69), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query 77 ELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPPGLLLNCCSSW 126
EL+ W +L DD P+ + + R+L V L +LK+ P L+ + C W
Sbjct 225 ELISKDWPELQDDI-PSILAQAQRILFVVDGLDELKVPPGALIQDICGDW 273
> cel:T12B5.13 fbxa-23; F-box A protein family member (fbxa-23)
Length=250
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 2/50 (4%)
Query 57 CARLLAILRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVC 106
CA+ L + LL E Q + L KW ++ D PT + +R +R+ C
Sbjct 150 CAKKLHLYDSLL--ENRQIVHLFHLKWFEIQLDSFPTNIATEIRDVRWNC 197
> sce:YNL273W TOF1; Subunit of a replication-pausing checkpoint
complex (Tof1p-Mrc1p-Csm3p) that acts at the stalled replication
fork to promote sister chromatid cohesion after DNA damage,
facilitating gap repair of damaged DNA; interacts with
the MCM helicase; K10997 replication fork protection complex
subunit Tof1/Swi1
Length=1238
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 47/86 (54%), Gaps = 2/86 (2%)
Query 1 AMAALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFV-AETNVQCMPCCAR 59
A LLN +S+ AQ+ D +F + RL + + R +L+ ++ + ++ ++Q M C
Sbjct 535 AFNELLNLVSRTKAAQEEDSTDIEFIVSRLFSDE-RIQLLSNLPKIGSKYSLQFMKSCIE 593
Query 60 LLAILRKLLKEETNQALELLQSKWRQ 85
L + K+L++ ++ +++ K R+
Sbjct 594 LTHSVLKVLEQYSDDKTLVIEGKSRR 619
> cel:F07A11.6 din-1; DAF-12 Interacting Protein family member
(din-1)
Length=2392
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Query 37 SELIMHILFVAETNVQCM-PCCARLLAILRKLLKEETNQALELLQSKWRQLADDCDPTKV 95
S + H+L A+ N Q M P +L A +++ N+A +++Q+K +Q + D K
Sbjct 1996 SGIFQHLLLHAQGNGQNMTPEMLQLKAAF--FAQQQENEANQMMQAKMKQQTINKDRIKE 2053
Query 96 EERVR 100
+ERV+
Sbjct 2054 QERVK 2058
> hsa:1821 DRP2, MGC133255; dystrophin related protein 2
Length=879
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query 1 AMAALLNRISQATTAQQIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARL 60
AM L +SQA + + D F++ L +L + V+ + A
Sbjct 161 AMEELSTTLSQAEGVRATWEPIGDLFIDSLPEHIQAIKLFKEEFSPMKDGVKLVNDLAHQ 220
Query 61 LAILRKLLKEETNQALELLQSKWRQLADDCDPTKVEERVRVLR 103
LAI L E +QALE + +W+QL V ER++ L+
Sbjct 221 LAISDVHLSMENSQALEQINVRWKQLQ-----ASVSERLKQLQ 258
> mmu:107476 Acaca, A530025K05Rik, Acac, Acc1, Gm738; acetyl-Coenzyme
A carboxylase alpha (EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2345
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 2/33 (6%)
Query 91 DPTKVEERVR--VLRFVCELIKLKLLPPGLLLN 121
DP+K+EE VR V+R+ L KL++L L +N
Sbjct 1483 DPSKIEESVRSMVMRYGSRLWKLRVLQAELKIN 1515
> hsa:31 ACACA, ACAC, ACC, ACC1, ACCA; acetyl-CoA carboxylase
alpha (EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA carboxylase /
biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2383
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 2/33 (6%)
Query 91 DPTKVEERVR--VLRFVCELIKLKLLPPGLLLN 121
DP+K+EE VR V+R+ L KL++L L +N
Sbjct 1521 DPSKIEESVRSMVMRYGSRLWKLRVLQAELKIN 1553
> dre:559403 acaca, im:7138837, si:ch211-199d18.1, wu:fj43d01;
acetyl-Coenzyme A carboxylase alpha; K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2349
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 2/33 (6%)
Query 91 DPTKVEERVR--VLRFVCELIKLKLLPPGLLLN 121
DP+K+EE VR V+R+ L KL++L L +N
Sbjct 1486 DPSKIEESVRSMVMRYGSRLWKLRVLQAELKIN 1518
> cel:F46B6.7 ztf-7; Zinc finger Transcription Factor family member
(ztf-7)
Length=614
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Query 6 LNRISQATTAQQIDQIAADFFLE----RLNAKQNRSELIMHILFVAETNV 51
+ R+ +A QQ ++ F L+ R NA+ NRS++I H+ + N+
Sbjct 284 MRRLEEALQCQQREREDTSFQLQCIFCRYNARGNRSKIIHHLYMIHHLNL 333
> cel:F11A10.4 mon-2; yeast MON (monensin-resistant) homolog family
member (mon-2)
Length=1646
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 1/61 (1%)
Query 22 AADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKE-ETNQALELLQ 80
A D + RL +EL H++ + + V MPCC + +L+K ++ Q L LLQ
Sbjct 1579 AVDSLIARLARDPKMTELYSHLVSLFPSVVDVMPCCHADPQLEHQLIKTIKSYQTLFLLQ 1638
Query 81 S 81
+
Sbjct 1639 N 1639
> tpv:TP01_0510 hypothetical protein
Length=228
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/34 (55%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Query 7 NRISQA-TTAQQIDQIAADFFLERL-NAKQNRSE 38
N++SQA QQI QI+ D F ERL NAK SE
Sbjct 153 NKLSQAFRIYQQIKQISKDEFTERLRNAKDEISE 186
> xla:414451 ube4a, MGC114906; ubiquitination factor E4A (UFD2
homolog); K10596 ubiquitin conjugation factor E4 A [EC:6.3.2.19]
Length=1072
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 26/56 (46%), Gaps = 6/56 (10%)
Query 71 ETNQALELLQSKWRQLADDCDPTKVEERVRVLR----FVCELIKLKLLPPGLLLNC 122
+ NQ+L LQS WR PT R + R ++C +K L P +L NC
Sbjct 532 KVNQSLHRLQSAWRDAQQSVSPTAENLREQFERLMTIYLC--LKTALSEPQMLQNC 585
> mmu:72978 Cnih3, 2900075G08Rik; cornichon homolog 3 (Drosophila)
Length=160
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 75 ALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLP 115
A + L++ ++ D C+P ER+R + +C L++ +LP
Sbjct 30 AFDELRTDFKSPIDQCNPVHARERLRNIERICFLLRKLVLP 70
> hsa:149111 CNIH3, CNIH-3, FLJ38993; cornichon homolog 3 (Drosophila)
Length=160
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 75 ALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLP 115
A + L++ ++ D C+P ER+R + +C L++ +LP
Sbjct 30 AFDELRTDFKSPIDQCNPVHARERLRNIERICFLLRKLVLP 70
> cpv:cgd5_670 reptin52; reptin like TIP49 family AAA+ ATpase
; K11338 RuvB-like protein 2 [EC:3.6.4.12]
Length=479
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 34/60 (56%), Gaps = 10/60 (16%)
Query 17 QIDQIAADFFLERLNAKQNRSELIMHILFVAETNVQCMPCCARLLAILRKLLKEETNQAL 76
QID+ A++ +++R+E++ +LF+ E ++ + C + L K L+EET+ L
Sbjct 270 QIDEKVAEW------KEESRAEIVHGVLFIDEVHMLDVEC----FSFLNKALEEETSPIL 319
> sce:YKL050C Protein of unknown function; the YKL050W protein
is a target of the SCFCdc4 ubiquitin ligase complex and YKL050W
transcription is regulated by Azf1p
Length=922
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 70 EETNQALELLQSKWRQLADDCDPTKVEERVRVLRFVCELIKLKLLPP 116
E + A L + + DDC T +E++ R F K+ L PP
Sbjct 106 ESVDLAARLASKRTKVSPDDCVETAIEQKARGEAFKVTFSKIPLTPP 152
Lambda K H
0.326 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40