bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1189_orf2
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
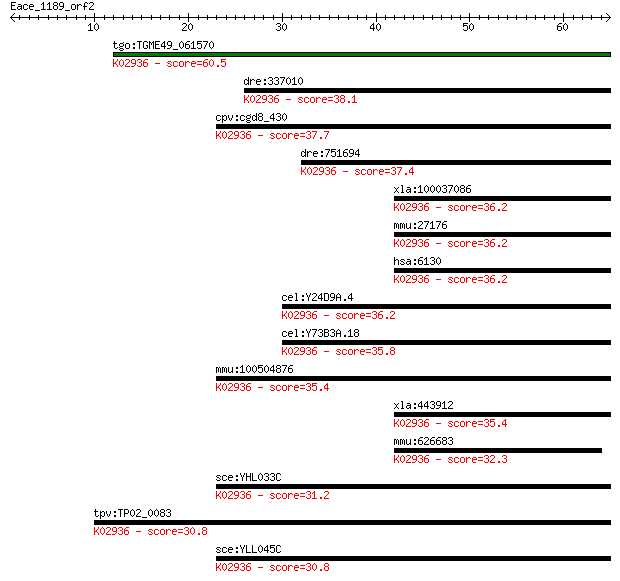
tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936... 60.5 1e-09
dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal ... 38.1 0.007
cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit ... 37.7 0.008
dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosom... 37.4 0.011
xla:100037086 hypothetical protein LOC100037086; K02936 large ... 36.2 0.027
mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K029... 36.2 0.027
hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 lar... 36.2 0.029
cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family m... 36.2 0.030
cel:Y73B3A.18 hypothetical protein; K02936 large subunit ribos... 35.8 0.031
mmu:100504876 60S ribosomal protein L7a-like; K02936 large sub... 35.4 0.044
xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a... 35.4 0.045
mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362; ... 32.3 0.34
sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosoma... 31.2 0.83
tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit ... 30.8 1.0
sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit r... 30.8 1.2
> tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936
large subunit ribosomal protein L7Ae
Length=276
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/54 (62%), Positives = 42/54 (77%), Gaps = 2/54 (3%)
Query 12 AAKKAAAGASKKKLPSNP-AGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
AA K ++G KKKLP+ P AGK+ KK KS IF KTPR+FR+G NIQPRR+L+R
Sbjct 9 AAAKGSSGG-KKKLPAAPLAGKSGKKVQKSPIFAKTPRSFRVGGNIQPRRDLSR 61
> dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal
protein L7a; K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 26 PSNPAGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
PS VKK L F+K P+NF IG +IQP+R+LTR
Sbjct 16 PSVAKKHEVKKVVNPL-FEKRPKNFGIGQDIQPKRDLTR 53
> cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit
ribosomal protein L7Ae
Length=263
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 31/46 (67%), Gaps = 4/46 (8%)
Query 23 KKLPSNPAGKNVKKTT----KSLIFQKTPRNFRIGCNIQPRRELTR 64
K + SN +G+N K T+ + + +KT RNFR+G +I+P+ +LTR
Sbjct 4 KMVASNKSGQNKKNTSTQKVRHPLIEKTSRNFRLGGDIRPKTDLTR 49
> dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosomal
protein L7Ae
Length=269
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 32 KNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
K V K ++ +F+K P+NF IG +IQP+R+L+R
Sbjct 23 KEVTKKVENPLFEKRPKNFGIGQDIQPKRDLSR 55
> xla:100037086 hypothetical protein LOC100037086; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF IG +IQP+R+LTR
Sbjct 31 LFEKRPKNFGIGQDIQPKRDLTR 53
> mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K02936
large subunit ribosomal protein L7Ae
Length=266
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF IG +IQP+R+LTR
Sbjct 31 LFEKRPKNFGIGQDIQPKRDLTR 53
> hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+NF IG +IQP+R+LTR
Sbjct 31 LFEKRPKNFGIGQDIQPKRDLTR 53
> cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family
member (rpl-7A); K02936 large subunit ribosomal protein L7Ae
Length=245
Score = 36.2 bits (82), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 30 AGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
A V+K K+ +F+K RNF IG +IQP++++TR
Sbjct 20 AQTQVQKEVKNPLFEKRARNFNIGQDIQPKKDVTR 54
> cel:Y73B3A.18 hypothetical protein; K02936 large subunit ribosomal
protein L7Ae
Length=234
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 30 AGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
A V+K K+ +F+K RNF IG +IQP++++TR
Sbjct 123 AQTQVQKEVKNPLFEKRARNFNIGQDIQPKKDVTR 157
> mmu:100504876 60S ribosomal protein L7a-like; K02936 large subunit
ribosomal protein L7Ae
Length=278
Score = 35.4 bits (80), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 30/46 (65%), Gaps = 4/46 (8%)
Query 23 KKLPSNPAGKNVKK-TTKSLI---FQKTPRNFRIGCNIQPRRELTR 64
KK+ PA VKK K+++ F+K P+NF I NIQP+R+LTR
Sbjct 20 KKVALAPAPATVKKQEAKNVVNPLFEKRPKNFSIKQNIQPKRDLTR 65
> xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELTR 64
+F+K P+N+ IG +IQP+R+LTR
Sbjct 31 LFEKRPKNYGIGQDIQPKRDLTR 53
> mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 42 IFQKTPRNFRIGCNIQPRRELT 63
+F+K P+NF IG +IQP+R LT
Sbjct 31 LFEKRPKNFGIGQDIQPKRVLT 52
> sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosomal
protein L7Ae
Length=256
Score = 31.2 bits (69), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 23 KKLPSNPAGKNVKKT--TKSLIFQKTPRNFRIGCNIQPRRELTR 64
KK+ P G K+ T++ + TP+NF IG +QP+R L+R
Sbjct 5 KKVAPAPFGAKSTKSNKTRNPLTHSTPKNFGIGQAVQPKRNLSR 48
> tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit
ribosomal protein L7Ae
Length=266
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 5/55 (9%)
Query 10 APAAKKAAAGASKKKLPSNPAGKNVKKTTKSLIFQKTPRNFRIGCNIQPRRELTR 64
AP KK SKK+ P K + K+ F++ PR+F IG +I+PR L+R
Sbjct 2 APTVKK-----SKKQPAPAPLAKPKETKAKNPFFERLPRDFGIGRSIRPRVNLSR 51
> sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit
ribosomal protein L7Ae
Length=256
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query 23 KKLPSNPAGKNVKKTTKSL--IFQKTPRNFRIGCNIQPRRELTR 64
KK+ P G K+ K+ + TP+NF IG +QP+R L+R
Sbjct 5 KKVAPAPFGAKSTKSNKAKNPLTHSTPKNFGIGQAVQPKRNLSR 48
Lambda K H
0.319 0.130 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2051595972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40