bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1196_orf2
Length=189
Score E
Sequences producing significant alignments: (Bits) Value
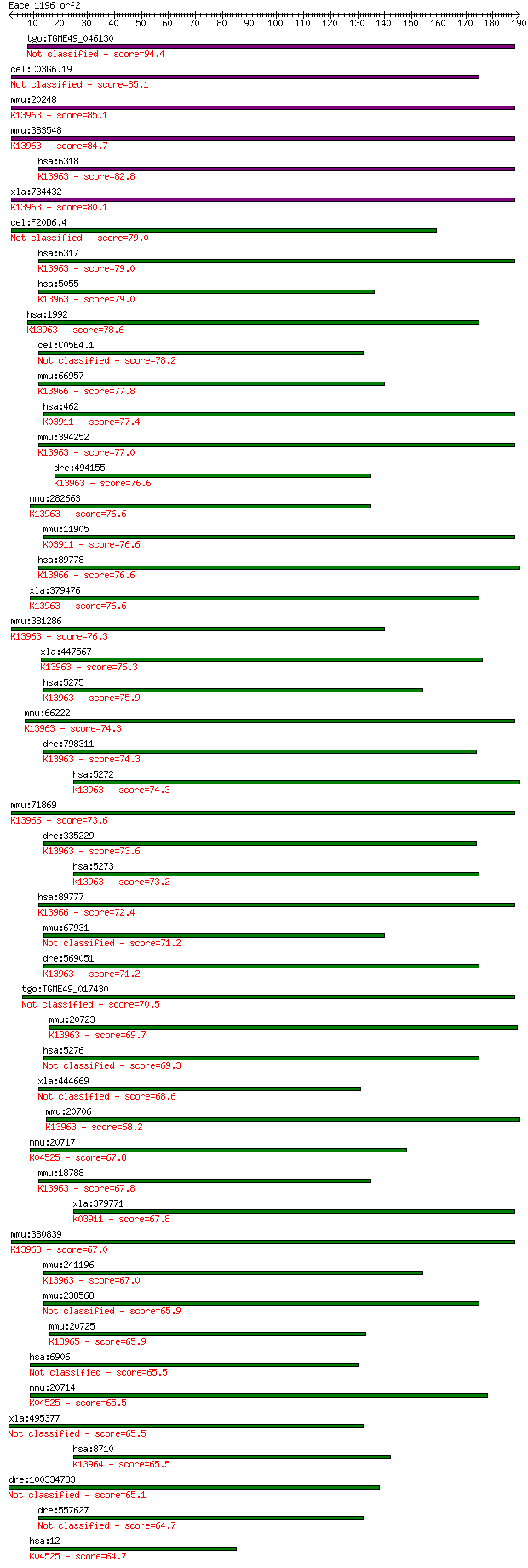
tgo:TGME49_046130 serine protease inhibitor, putative (EC:2.7.... 94.4 2e-19
cel:C03G6.19 srp-6; SeRPin family member (srp-6) 85.1 1e-16
mmu:20248 Serpinb3a, 1110001H02Rik, 1110013A16Rik, Scca2, Serp... 85.1 1e-16
mmu:383548 Serpinb3b, MGC118408, Scca2-rs, Scca2-rs1; serine (... 84.7 2e-16
hsa:6318 SERPINB4, LEUPIN, PI11, SCCA-2, SCCA1, SCCA2; serpin ... 82.8 6e-16
xla:734432 serpinb1, MGC115155, elanh2, lei, m/nei, mnei, pi2;... 80.1 5e-15
cel:F20D6.4 srp-7; SeRPin family member (srp-7) 79.0 8e-15
hsa:6317 SERPINB3, HsT1196, SCC, SCCA-1, SCCA-PD, SCCA1, T4-A;... 79.0 9e-15
hsa:5055 SERPINB2, HsT1201, PAI, PAI-2, PAI2, PLANH2; serpin p... 79.0 1e-14
hsa:1992 SERPINB1, EI, ELANH2, LEI, M/NEI, MNEI, PI2; serpin p... 78.6 1e-14
cel:C05E4.1 srp-2; SeRPin family member (srp-2) 78.2 2e-14
mmu:66957 Serpinb11, 2310046M08Rik, AU015399; serine (or cyste... 77.8 2e-14
hsa:462 SERPINC1, AT3, ATIII, MGC22579; serpin peptidase inhib... 77.4 3e-14
mmu:394252 Serpinb3d, Serpinb3l1; serine (or cysteine) peptida... 77.0 4e-14
dre:494155 serpinb1l1, serpinb1; serpin peptidase inhibitor, c... 76.6 4e-14
mmu:282663 Serpinb1b, 6330533H24Rik, EIB, Serpin1b1, ovalbumin... 76.6 5e-14
mmu:11905 Serpinc1, AI114908, ATIII, At-3, At3; serine (or cys... 76.6 5e-14
hsa:89778 SERPINB11, EPIPIN, MGC163242, MGC163244, SERPIN11; s... 76.6 5e-14
xla:379476 serpinb6, MGC64421; serpin peptidase inhibitor, cla... 76.6 5e-14
mmu:381286 Serpinb3c, 1110001H02Rik, 1110013A16Rik, Scca2, Ser... 76.3 6e-14
xla:447567 serpinb4, MGC84275; serpin peptidase inhibitor, cla... 76.3 7e-14
hsa:5275 SERPINB13, HSHUR7SEQ, HUR7, MGC126870, PI13, headpin;... 75.9 7e-14
mmu:66222 Serpinb1a, 1190005M04Rik, AI325983, EI, EIA, ELANH2,... 74.3 2e-13
dre:798311 zgc:173729; K13963 serpin B 74.3 2e-13
hsa:5272 SERPINB9, CAP-3, CAP3, PI9; serpin peptidase inhibito... 74.3 2e-13
mmu:71869 Serpinb12, 2300003F07Rik, 4833409F13Rik; serine (or ... 73.6 4e-13
dre:335229 serpinb1l4, fk92d08, wu:fk92d08, zgc:77645; serpin ... 73.6 4e-13
hsa:5273 SERPINB10, PI10, bomapin; serpin peptidase inhibitor,... 73.2 6e-13
hsa:89777 SERPINB12, MGC119247, MGC119248, YUKOPIN; serpin pep... 72.4 8e-13
mmu:67931 Serpini2, 1810006A24Rik, Spi14; serine (or cysteine)... 71.2 2e-12
dre:569051 im:7148243; K13963 serpin B 71.2 2e-12
tgo:TGME49_017430 protease inhibitor, putative (EC:2.7.11.1) 70.5 4e-12
mmu:20723 Serpinb9, BB283241, CAP-3, CAP3, PI-9, PI9, Spi6, ov... 69.7 6e-12
hsa:5276 SERPINI2, MEPI, PANCPIN, PI14, TSA2004; serpin peptid... 69.3 7e-12
xla:444669 serpini1, MGC84260, neuroserpin; serpin peptidase i... 68.6 1e-11
mmu:20706 Serpinb9b, 1600019A21Rik, R86, SPI-CI, Spi10, ovalbu... 68.2 2e-11
mmu:20717 Serpina3m, 3e46, AI195004, MMCM7, MMSPi2.4, Spi-2l, ... 67.8 2e-11
mmu:18788 Serpinb2, PAI-2, Planh2, ovalbumin; serine (or cyste... 67.8 2e-11
xla:379771 serpinc1, MGC52682; serpin peptidase inhibitor, cla... 67.8 2e-11
mmu:380839 Serpinb1c, EIC, MGC129308, ovalbumin; serine (or cy... 67.0 3e-11
mmu:241196 Serpinb13, 5430417G24, HUR7, HURPIN, PI13, headpin;... 67.0 4e-11
mmu:238568 Serpinb6d, Gm11390, OTTMUSG00000000712, SPI3D; seri... 65.9 8e-11
mmu:20725 Serpinb8, CAP-2, CAP2, NK10, Spi8, ovalbumin; serine... 65.9 9e-11
hsa:6906 SERPINA7, FLJ17820, TBG; serpin peptidase inhibitor, ... 65.5 1e-10
mmu:20714 Serpina3k, 1300001I07Rik, D12Rp54, MMCM2, MMSpi2, RP... 65.5 1e-10
xla:495377 serpina3; serpin peptidase inhibitor, clade A (alph... 65.5 1e-10
hsa:8710 SERPINB7, DKFZp686D06190, MEGSIN, MGC120014, MGC12001... 65.5 1e-10
dre:100334733 hypothetical protein LOC100334733 65.1 2e-10
dre:557627 serpini1, serpini1l, si:ch211-167c22.4; serpin pept... 64.7 2e-10
hsa:12 SERPINA3, AACT, ACT, GIG25, MGC88254; serpin peptidase ... 64.7 2e-10
> tgo:TGME49_046130 serine protease inhibitor, putative (EC:2.7.11.1)
Length=432
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 60/193 (31%), Positives = 99/193 (51%), Gaps = 17/193 (8%)
Query 8 CDAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASP 67
+A + DF D+AAA IN FV +T HIK +V A + P++++VLV+A+YFK WA
Sbjct 137 TEAKTLDFADTAAAVEEINGFVSKQTHEHIKQLVTAQDVNPNTKLVLVSAMYFKCSWAKQ 196
Query 68 FSPHRTATGPFY--VNDTRTGRSEVQQVKFMQQRLEED-FGLFESDKVKVLSMPYADPRC 124
F HRT TG F+ VN G+ QQV M L++ + + V +++PY+DP
Sbjct 197 FPKHRTDTGTFHALVN----GKHVEQQVSMMHTTLKDSPLAVKVDENVVAIALPYSDPNT 252
Query 125 RLYLFLPNSIAAFEQELQQKPET------TEELVSLVDNTPSSDVV----LDLSLPLIKL 174
+Y+ P +K E LV + + +++ + + L++P KL
Sbjct 253 AMYIIQPRDSHHLATLFDKKKSAELGVAYIESLVREMRSEETAEAMWGKQVRLTMPKFKL 312
Query 175 AAERNQVGLVDVY 187
++ N+ L+ V+
Sbjct 313 SSAANREDLIPVF 325
> cel:C03G6.19 srp-6; SeRPin family member (srp-6)
Length=375
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 55/173 (31%), Positives = 86/173 (49%), Gaps = 10/173 (5%)
Query 2 VSERLNCDAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFK 61
V + N A +FED A+A AIN FV TKGHIK I+ +++ VL NA YFK
Sbjct 115 VKKLYNAGASQLNFEDQEASAEAINNFVSENTKGHIKKIINPDSISEELVAVLTNAFYFK 174
Query 62 APWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYAD 121
A W + F T F+ ++ +E + ++ E+ G F+ VLS+PY D
Sbjct 175 ANWQTKFKKESTYKREFFSSENSKRETEFLHSRNSNRKYSEN-GQFQ-----VLSLPYKD 228
Query 122 PRCRLYLFLPNSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKL 174
L +FLP + + LQ T + L+ NT S+ ++++++P K+
Sbjct 229 TSFALSIFLPKTRFGLSEALQNLDSVT--IQQLMSNT--SNTLVNIAMPKWKI 277
> mmu:20248 Serpinb3a, 1110001H02Rik, 1110013A16Rik, Scca2, Serpinb4,
Sqn5; serine (or cysteine) peptidase inhibitor, clade
B (ovalbumin), member 3A; K13963 serpin B
Length=387
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 56/193 (29%), Positives = 93/193 (48%), Gaps = 14/193 (7%)
Query 2 VSERLNCDAVSTDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
+ E + S DFE +A + IN++VE +T G IK + P +L S+ MVLVNA+YF
Sbjct 120 IKEYYQANVESLDFEHAAEESEKKINSWVESQTNGKIKDLFPNGSLNRSTIMVLVNAVYF 179
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYA 120
K W F T F++N ++ + V+ M+Q +E +F E + K++ +PY
Sbjct 180 KGQWNHKFDEKHTTEEKFWLN-----KNTSKPVQMMKQNIEFNFMFLEDVQAKIVEIPYK 234
Query 121 DPRCRLYLFLPNSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN- 179
+ + LP I +Q +Q T ++L+ L LSLP K+ + +
Sbjct 235 GKELSMIVLLPVEINGLKQLEEQL--TADKLLEWTRAENMHMTELYLSLPRFKVDEKYDL 292
Query 180 -----QVGLVDVY 187
+G+VD +
Sbjct 293 PIPLEHMGMVDAF 305
> mmu:383548 Serpinb3b, MGC118408, Scca2-rs, Scca2-rs1; serine
(or cysteine) peptidase inhibitor, clade B (ovalbumin), member
3B; K13963 serpin B
Length=387
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 54/193 (27%), Positives = 96/193 (49%), Gaps = 14/193 (7%)
Query 2 VSERLNCDAVSTDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
+ E S DFE + + IN++VE KT G IK + P+ +L+ S+ +VLVNA+YF
Sbjct 120 IKEYYQAKVESLDFEHATEESEKKINSWVESKTNGKIKDLFPSGSLSSSTILVLVNAVYF 179
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYA 120
K W F+ + T F++N ++ + V+ M+QR + +F +++ +PY
Sbjct 180 KGQWNRKFNENHTREEKFWLN-----KNTSKPVQMMKQRNKFNFSFLGDVHAQIVEIPYK 234
Query 121 DPRCRLYLFLPNSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN- 179
+++ LP I +Q +Q TT++L+ + L LSLP K+ + +
Sbjct 235 GKDLSMFVLLPMEIDGLKQLEEQL--TTDKLLEWIKAENMHLTELYLSLPRFKVEEKYDL 292
Query 180 -----QVGLVDVY 187
+G+VD +
Sbjct 293 QVPLEHMGMVDAF 305
> hsa:6318 SERPINB4, LEUPIN, PI11, SCCA-2, SCCA1, SCCA2; serpin
peptidase inhibitor, clade B (ovalbumin), member 4; K13963
serpin B
Length=390
Score = 82.8 bits (203), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 53/183 (28%), Positives = 91/183 (49%), Gaps = 14/183 (7%)
Query 12 STDFEDS-AAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
STDF ++ + IN++VE +T IK++ P + + +VLVNA+YFK W + F
Sbjct 133 STDFANAPEESRKKINSWVESQTNEKIKNLFPDGTIGNDTTLVLVNAIYFKGQWENKFKK 192
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T F+ N ++ + V+ M+Q +F L E + KVL +PY + + L
Sbjct 193 ENTKEEKFWPN-----KNTYKSVQMMRQYNSFNFALLEDVQAKVLEIPYKGKDLSMIVLL 247
Query 131 PNSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN------QVGLV 184
PN I Q+L++K T E+L+ + +DL LP K+ + +G+V
Sbjct 248 PNEIDGL-QKLEEKL-TAEKLMEWTSLQNMRETCVDLHLPRFKMEESYDLKDTLRTMGMV 305
Query 185 DVY 187
+++
Sbjct 306 NIF 308
> xla:734432 serpinb1, MGC115155, elanh2, lei, m/nei, mnei, pi2;
serpin peptidase inhibitor, clade B (ovalbumin), member 1;
K13963 serpin B
Length=377
Score = 80.1 bits (196), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 56/195 (28%), Positives = 95/195 (48%), Gaps = 16/195 (8%)
Query 2 VSERLNCDAVSTDFEDSAA-AAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
V ++ + D + DF + A IN +V +TKG I ++ + S+++VLVNA+YF
Sbjct 109 VKKQYSADLGTVDFISALEDARKEINKWVSEQTKGKIPEVLSTGTVDRSTKLVLVNAIYF 168
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYA 120
K WA F+ T PF +N + E + VK M Q+ + F ++L +PY
Sbjct 169 KGDWAKKFNAEHTTDMPFQLN-----KKEQKTVKMMYQKEKLPFNYIPDINCRILELPYV 223
Query 121 DPRCRLYLFLPNSIAAFEQELQQ--KPETTEELVSLVDNTPSSDVVLDLSLPLI------ 172
D + + LP++I LQQ K + E++ +N +DV + LP
Sbjct 224 DYELSMIIILPDNINDDTTGLQQLEKELSLEKIHEWTENMMPTDV--HIHLPKFKLEDSY 281
Query 173 KLAAERNQVGLVDVY 187
KL ++ +G+VD++
Sbjct 282 KLKSQLAGMGMVDLF 296
> cel:F20D6.4 srp-7; SeRPin family member (srp-7)
Length=373
Score = 79.0 bits (193), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 51/159 (32%), Positives = 78/159 (49%), Gaps = 9/159 (5%)
Query 2 VSERLNCDAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFK 61
V + N A S DF++ A A AIN FV T HIK I+ + ++ VL NALYFK
Sbjct 113 VKKLYNAGASSLDFDNKEATAEAINNFVRENTGDHIKKIIGSDSINSDLVAVLTNALYFK 172
Query 62 APWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQ-QRLEEDFGLFESDKVKVLSMPYA 120
A W + F T F+ + +++ F+ + D+ E+D+ +VLS+PY
Sbjct 173 ADWQNKFKKDSTFKSEFF-----SSADSKREIDFLHASSVSRDYA--ENDQFQVLSLPYK 225
Query 121 DPRCRLYLFLPNSIAAFEQELQQ-KPETTEELVSLVDNT 158
D L +FLP + + L+ T + L+S V +T
Sbjct 226 DNTFALTIFLPKTRFGLTESLKTLDSATIQHLLSNVSST 264
> hsa:6317 SERPINB3, HsT1196, SCC, SCCA-1, SCCA-PD, SCCA1, T4-A;
serpin peptidase inhibitor, clade B (ovalbumin), member 3;
K13963 serpin B
Length=390
Score = 79.0 bits (193), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 51/183 (27%), Positives = 90/183 (49%), Gaps = 14/183 (7%)
Query 12 STDFEDS-AAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
S DF ++ + IN++VE +T IK+++P + ++ +VLVNA+YFK W F+
Sbjct 133 SVDFANAPEESRKKINSWVESQTNEKIKNLIPEGNIGSNTTLVLVNAIYFKGQWEKKFNK 192
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T F+ N ++ + ++ M+Q F E + KVL +PY + + L
Sbjct 193 EDTKEEKFWPN-----KNTYKSIQMMRQYTSFHFASLEDVQAKVLEIPYKGKDLSMIVLL 247
Query 131 PNSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN------QVGLV 184
PN I Q+L++K T E+L+ + +DL LP K+ + +G+V
Sbjct 248 PNEIDGL-QKLEEKL-TAEKLMEWTSLQNMRETRVDLHLPRFKVEESYDLKDTLRTMGMV 305
Query 185 DVY 187
D++
Sbjct 306 DIF 308
> hsa:5055 SERPINB2, HsT1201, PAI, PAI-2, PAI2, PLANH2; serpin
peptidase inhibitor, clade B (ovalbumin), member 2; K13963
serpin B
Length=415
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 74/125 (59%), Gaps = 7/125 (5%)
Query 12 STDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
+ DF + A A IN++V+ +TKG I +++P ++ +RMVLVNA+YFK W +PF
Sbjct 155 AVDFLECAEEARKKINSWVKTQTKGKIPNLLPEGSVDGDTRMVLVNAVYFKGKWKTPFEK 214
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
PF VN + R+ VQ + +++++L + G E K ++L +PYA ++L L
Sbjct 215 KLNGLYPFRVNSAQ--RTPVQMM-YLREKL--NIGYIEDLKAQILELPYAGD-VSMFLLL 268
Query 131 PNSIA 135
P+ IA
Sbjct 269 PDEIA 273
> hsa:1992 SERPINB1, EI, ELANH2, LEI, M/NEI, MNEI, PI2; serpin
peptidase inhibitor, clade B (ovalbumin), member 1; K13963
serpin B
Length=379
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 51/170 (30%), Positives = 83/170 (48%), Gaps = 8/170 (4%)
Query 8 CDAVSTDFED-SAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWAS 66
D S DF+ S A IN +V+G+T+G I ++ + + +++VLVNA+YFK W
Sbjct 114 ADLASVDFQHASEDARKTINQWVKGQTEGKIPELLASGMVDNMTKLVLVNAIYFKGNWKD 173
Query 67 PFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRL 126
F T PF +N + + + VK M Q+ + +G E K +VL +PY +
Sbjct 174 KFMKEATTNAPFRLN-----KKDRKTVKMMYQKKKFAYGYIEDLKCRVLELPYQGEELSM 228
Query 127 YLFLPNSIAAFEQELQQKPE--TTEELVSLVDNTPSSDVVLDLSLPLIKL 174
+ LP+ I L++ E T E+L + +++SLP KL
Sbjct 229 VILLPDDIEDESTGLKKIEEQLTLEKLHEWTKPENLDFIEVNVSLPRFKL 278
> cel:C05E4.1 srp-2; SeRPin family member (srp-2)
Length=359
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 46/120 (38%), Positives = 66/120 (55%), Gaps = 7/120 (5%)
Query 12 STDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPH 71
S DF + AA +N FVE T G IK ++PA + ++ LVNA+YFKA W S F+
Sbjct 112 SLDFSQTEQAAKTMNTFVENHTNGKIKDLIPADS-ANNAFAFLVNAMYFKADWQSKFAKE 170
Query 72 RTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLP 131
T F+ T +E +Q+ F+ + L+E E +VLS+ YADP+ L +FLP
Sbjct 171 STTGREFF-----TSEAESRQIPFLTE-LDEHRDYTEDVLFQVLSLKYADPKFTLAIFLP 224
> mmu:66957 Serpinb11, 2310046M08Rik, AU015399; serine (or cysteine)
peptidase inhibitor, clade B (ovalbumin), member 11;
K13966 serpin B11/12
Length=388
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 68/129 (52%), Gaps = 6/129 (4%)
Query 12 STDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
+ DFE S +INA+V+ KT G I ++ + PSS MVLV+A+YFK W + F
Sbjct 132 TVDFELSTEETRKSINAWVKNKTNGKITNLFAKGTIDPSSVMVLVSAIYFKGQWQNKFQK 191
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T PF++ G+S V V M Q + + +++VL +PYA+ + R+ + L
Sbjct 192 RETVKAPFHMG---VGKSAV--VNMMYQTGTFKLAIIKEPEMQVLELPYANNKLRMIILL 246
Query 131 PNSIAAFEQ 139
P A+ Q
Sbjct 247 PVGTASVSQ 255
> hsa:462 SERPINC1, AT3, ATIII, MGC22579; serpin peptidase inhibitor,
clade C (antithrombin), member 1; K03911 antithrombin
III
Length=464
Score = 77.4 bits (189), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 56/186 (30%), Positives = 95/186 (51%), Gaps = 27/186 (14%)
Query 14 DFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF+++A + AAIN +V KT+G I ++P+ A+ + +VLVN +YFK W S FSP
Sbjct 206 DFKENAEQSRAAINKWVSNKTEGRITDVIPSEAINELTVLVLVNTIYFKGLWKSKFSPEN 265
Query 73 TATGPFYVNDTRTGRSEV--QQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T FY D + + + Q+ KF +R+ E +VL +P+ + L L
Sbjct 266 TRKELFYKADGESCSASMMYQEGKFRYRRVAEG--------TQVLELPFKGDDITMVLIL 317
Query 131 P---NSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIK------LAAERNQV 181
P S+A E+EL PE +E + ++ +++L + +P + L + +
Sbjct 318 PKPEKSLAKVEKELT--PEVLQEWLDELE-----EMMLVVHMPRFRIEDGFSLKEQLQDM 370
Query 182 GLVDVY 187
GLVD++
Sbjct 371 GLVDLF 376
> mmu:394252 Serpinb3d, Serpinb3l1; serine (or cysteine) peptidase
inhibitor, clade B (ovalbumin), member 3D; K13963 serpin
B
Length=387
Score = 77.0 bits (188), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 51/186 (27%), Positives = 93/186 (50%), Gaps = 20/186 (10%)
Query 12 STDFEDSAA-AAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
S DF +A + IN+++ +T G IK + P+ +L S+ +VLVNA+YFK W F
Sbjct 130 SLDFAHAAEESQKKINSWMARQTNGKIKDLFPSGSLNSSTILVLVNAVYFKGQWNHKFDE 189
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T F++N ++ + V+ M+QR + +F E+ + K++ +PY +++ L
Sbjct 190 KHTREEKFWLN-----KNTSKPVQMMKQRNKFNFIFLENVQAKIVEIPYKGKELSMFVLL 244
Query 131 P---NSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN------QV 181
P + + FE++L T ++L+ L LSLP K+ + + +
Sbjct 245 PVEIDGLKKFEEQL-----TADKLLQWTRAENMHMTELYLSLPQFKVEEKYDLRVPLEHM 299
Query 182 GLVDVY 187
G+VD +
Sbjct 300 GMVDAF 305
> dre:494155 serpinb1l1, serpinb1; serpin peptidase inhibitor,
clade B (ovalbumin), member 1, like 1; K13963 serpin B
Length=384
Score = 76.6 bits (187), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/117 (36%), Positives = 60/117 (51%), Gaps = 5/117 (4%)
Query 18 SAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTATGP 77
S A IN +VE T+ IK ++P+ A+ +R+VLVNA+YFK W F T G
Sbjct 130 SEDARVNINTWVEKNTQEKIKDLLPSGAIDAMTRLVLVNAIYFKGNWEEKFPKEATRDGV 189
Query 78 FYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPNSI 134
F +N +T + VK M Q+ E G E K VL +PYA + + LP+ I
Sbjct 190 FRLNKNQT-----KPVKMMHQKAEFPSGYIEEMKSHVLELPYAGKNLSMLIILPDEI 241
> mmu:282663 Serpinb1b, 6330533H24Rik, EIB, Serpin1b1, ovalbumin;
serine (or cysteine) peptidase inhibitor, clade B, member
1b; K13963 serpin B
Length=382
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 41/127 (32%), Positives = 64/127 (50%), Gaps = 6/127 (4%)
Query 9 DAVSTDFEDSAA-AAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASP 67
D + DF+ ++ A IN +V+G+T+G I ++ + +++VLVNA+YFK W
Sbjct 115 DLAAVDFQHASEDARKEINQWVKGQTEGKIPELLAKGVVDSMTKLVLVNAIYFKGIWEEQ 174
Query 68 FSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLY 127
F T PF +N + + + VK M Q+ + FG K KVL MPY +
Sbjct 175 FMTRETINAPFRLN-----KKDTKTVKMMYQKKKFPFGYISDLKCKVLEMPYQGGELSMV 229
Query 128 LFLPNSI 134
+ LP I
Sbjct 230 ILLPEDI 236
> mmu:11905 Serpinc1, AI114908, ATIII, At-3, At3; serine (or cysteine)
peptidase inhibitor, clade C (antithrombin), member
1; K03911 antithrombin III
Length=465
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 57/186 (30%), Positives = 91/186 (48%), Gaps = 27/186 (14%)
Query 14 DF-EDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF E+ + IN +V KT+G IK ++P A+ + +VLVN +YFK W S FSP
Sbjct 207 DFKENPEQSRVTINNWVANKTEGRIKDVIPQGAINELTALVLVNTIYFKGLWKSKFSPEN 266
Query 73 TATGPFYVNDTRTGRSEV--QQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T PFY D ++ + Q+ KF +R+ E +VL +P+ + L L
Sbjct 267 TRKEPFYKVDGQSCPVPMMYQEGKFKYRRVAEG--------TQVLELPFKGDDITMVLIL 318
Query 131 P---NSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIK------LAAERNQV 181
P S+A EQEL PE +E + + S+ +L + +P + L + +
Sbjct 319 PKPEKSLAKVEQELT--PELLQEWLDEL-----SETMLVVHMPRFRTEDGFSLKEQLQDM 371
Query 182 GLVDVY 187
GL+D++
Sbjct 372 GLIDLF 377
> hsa:89778 SERPINB11, EPIPIN, MGC163242, MGC163244, SERPIN11;
serpin peptidase inhibitor, clade B (ovalbumin), member 11
(gene/pseudogene); K13966 serpin B11/12
Length=392
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 55/189 (29%), Positives = 90/189 (47%), Gaps = 22/189 (11%)
Query 12 STDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
+ DFE S INA+VE KT G + ++ S + PSS MVLVNA+YFK W + F
Sbjct 135 TVDFEQSTEETRKTINAWVENKTNGKVANLFGKSTIDPSSVMVLVNAIYFKGQWQNKFQV 194
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T PF +++ + V+ M Q + +++VL +PY + + + + L
Sbjct 195 RETVKSPFQLSEGKN-----VTVEMMYQIGTFKLAFVKEPQMQVLELPYVNNKLSMIILL 249
Query 131 PNSIAAFEQ-ELQQKPETTEELVSLVDNTPSSDVV---LDLSLPLIKLAAER------NQ 180
P IA +Q E Q T E T SS+++ +++ LP KL +
Sbjct 250 PVGIANLKQIEKQLNSGTFHEW------TSSSNMMEREVEVHLPRFKLETKYELNSLLKS 303
Query 181 VGLVDVYKR 189
+G+ D++ +
Sbjct 304 LGVTDLFNQ 312
> xla:379476 serpinb6, MGC64421; serpin peptidase inhibitor, clade
B (ovalbumin), member 6; K13963 serpin B
Length=379
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 53/169 (31%), Positives = 84/169 (49%), Gaps = 8/169 (4%)
Query 9 DAVSTDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASP 67
D + DF A + IN +V KT+G I ++P A+ +R+VLVNA+YFK WA+
Sbjct 115 DLRAVDFCKKAEESRGEINEWVAQKTEGKINDLLPVGAVDSLTRLVLVNAIYFKGNWANK 174
Query 68 FSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLY 127
F+P T PF +N ++E + V+ M ++ + KV+ +PY D +
Sbjct 175 FNPEHTHEMPFRLN-----KNETKPVQMMYKKAKFPMTYVGELFTKVIEIPYVDNELSMI 229
Query 128 LFLPNSIAAFEQELQQ-KPETTEELVSLVDNTPSSDVV-LDLSLPLIKL 174
+ LP+ I L++ + E T E N D+ ++LSLP KL
Sbjct 230 ILLPDDINDGTTGLEKLEKELTYEKFLQWTNPEMMDITEMELSLPKFKL 278
> mmu:381286 Serpinb3c, 1110001H02Rik, 1110013A16Rik, Scca2, Serpinb4;
serine (or cysteine) peptidase inhibitor, clade B,
member 3C; K13963 serpin B
Length=386
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 73/139 (52%), Gaps = 6/139 (4%)
Query 2 VSERLNCDAVSTDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
+ E + + S DFE +A + IN +V+ +T G IK + P+ +L+ S+++VLVNA+YF
Sbjct 120 IKEYYHANVESLDFEHAAEESEKKINFWVKNETNGKIKDLFPSGSLSSSTKLVLVNAVYF 179
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYA 120
K W F + T F++N ++ V M+QR + F E + +++ +PY
Sbjct 180 KGRWNHKFDENNTIEEMFWLN-----KNTSIPVPMMKQRNKFMFSFLEDVQAQIVEIPYK 234
Query 121 DPRCRLYLFLPNSIAAFEQ 139
+++ LP I +Q
Sbjct 235 GKELSMFVLLPMEIDGLKQ 253
> xla:447567 serpinb4, MGC84275; serpin peptidase inhibitor, clade
B (ovalbumin), member 4; K13963 serpin B
Length=392
Score = 76.3 bits (186), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 57/169 (33%), Positives = 83/169 (49%), Gaps = 19/169 (11%)
Query 13 TDFEDSA-AAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPH 71
TDF+++A A+ INA+VE KTK IK++ P L S+ + LVNA+YFK W F
Sbjct 136 TDFKNNAEASIVKINAWVECKTKSKIKNLFPKGTLDDSTVLALVNAVYFKGRWKKQFEKE 195
Query 72 RTATGPFYV--NDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLF 129
T PF++ NDT T VK M Q + G K ++L +PY + + +
Sbjct 196 NTKDAPFFLKTNDTTT-------VKMMSQTGKYKLGSHPELKCRILELPYEEG-FSMKII 247
Query 130 LPNSI---AAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLA 175
LP+ I A E L T E+L L++ +V + + LP K
Sbjct 248 LPDDIDGLAELEANL-----TYEKLTKLMNLENIREVKVVVRLPQFKFG 291
> hsa:5275 SERPINB13, HSHUR7SEQ, HUR7, MGC126870, PI13, headpin;
serpin peptidase inhibitor, clade B (ovalbumin), member 13;
K13963 serpin B
Length=391
Score = 75.9 bits (185), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 72/142 (50%), Gaps = 7/142 (4%)
Query 14 DFEDSA-AAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF ++A + IN++VE KT IK + P +++ S+++VLVN +YFK W F
Sbjct 136 DFVNAADESRKKINSWVESKTNEKIKDLFPDGSISSSTKLVLVNMVYFKGQWDREFKKEN 195
Query 73 TATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
T F++N +S + V+ M Q F E + K+L +PY + +++ LPN
Sbjct 196 TKEEKFWMN-----KSTSKSVQMMTQSHSFSFTFLEDLQAKILGIPYKNNDLSMFVLLPN 250
Query 133 SIAAFEQELQQ-KPETTEELVS 153
I E+ + + PE E S
Sbjct 251 DIDGLEKIIDKISPEKLVEWTS 272
> mmu:66222 Serpinb1a, 1190005M04Rik, AI325983, EI, EIA, ELANH2,
LEI, M/NEI, MGC129309, MNEI, PI2; serine (or cysteine) peptidase
inhibitor, clade B, member 1a; K13963 serpin B
Length=379
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 53/190 (27%), Positives = 84/190 (44%), Gaps = 14/190 (7%)
Query 7 NCDAVSTDF-EDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWA 65
D DF S A IN +V+G+T+G I ++ + +++VLVNA+YFK W
Sbjct 113 GADLAPVDFLHASEDARKEINQWVKGQTEGKIPELLSVGVVDSMTKLVLVNAIYFKGMWE 172
Query 66 SPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCR 125
F T PF R + + + VK M Q+ + FG K KVL MPY
Sbjct 173 EKFMTEDTTDAPF-----RLSKKDTKTVKMMYQKKKFPFGYISDLKCKVLEMPYQGGELS 227
Query 126 LYLFLPNSIAAFEQELQ--QKPETTEELVSLVDNTPSSDVVLDLSLPLIK------LAAE 177
+ + LP I L+ +K T E+L+ + + + LP K L +
Sbjct 228 MVILLPKDIEDESTGLKKIEKQITLEKLLEWTKRENLEFIDVHVKLPRFKIEESYTLNSN 287
Query 178 RNQVGLVDVY 187
++G+ D++
Sbjct 288 LGRLGVQDLF 297
> dre:798311 zgc:173729; K13963 serpin B
Length=439
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 51/163 (31%), Positives = 79/163 (48%), Gaps = 8/163 (4%)
Query 14 DFED-SAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF++ S A+ IN +VE T+ IK ++P+ A+ +R+VLVNA+YFK W F+
Sbjct 178 DFKNKSEASRVNINKWVEKNTQEKIKDLLPSGAIDAMTRLVLVNAIYFKGNWEKKFTKEA 237
Query 73 TATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
T G F +N +T + VK M Q+ +VL +PYA + + LP+
Sbjct 238 TRDGQFKLNKNQT-----KPVKMMHQKARFSLASIPEMNSQVLELPYAGKNLSMLIILPD 292
Query 133 SIAAFEQELQ--QKPETTEELVSLVDNTPSSDVVLDLSLPLIK 173
I LQ +K T E+L+ + + +SLP K
Sbjct 293 QIEDATTGLQKLEKALTYEKLMEWTKPSMMCQQEVQVSLPKFK 335
> hsa:5272 SERPINB9, CAP-3, CAP3, PI9; serpin peptidase inhibitor,
clade B (ovalbumin), member 9; K13963 serpin B
Length=376
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 52/173 (30%), Positives = 85/173 (49%), Gaps = 17/173 (9%)
Query 25 INAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTATGPFYVNDTR 84
IN +V KT+G I+ ++P S++ +R+VLVNA+YFK W PF T PF +N
Sbjct 132 INTWVSKKTEGKIEELLPGSSIDAETRLVLVNAIYFKGKWNEPFDETYTREMPFKIN--- 188
Query 85 TGRSEVQQVKFMQQRLEEDFGLFESDKVK--VLSMPYADPRCRLYLFLPNSIAAFEQELQ 142
+ E + V+ M Q E F L +V+ +L +PYA L + LP+ E
Sbjct 189 --QEEQRPVQMMYQ--EATFKLAHVGEVRAQLLELPYARKELSLLVLLPDD--GVELSTV 242
Query 143 QKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN------QVGLVDVYKR 189
+K T E+L + +++ LP KL + + +G+VD +++
Sbjct 243 EKSLTFEKLTAWTKPDCMKSTEVEVLLPKFKLQEDYDMESVLRHLGIVDAFQQ 295
> mmu:71869 Serpinb12, 2300003F07Rik, 4833409F13Rik; serine (or
cysteine) peptidase inhibitor, clade B (ovalbumin), member
12; K13966 serpin B11/12
Length=423
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 57/200 (28%), Positives = 98/200 (49%), Gaps = 24/200 (12%)
Query 2 VSERLNCDAVSTDFE-DSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
V+E + S DF+ DS + IN +VE +++G IK + A+ S+ +VLVNA+YF
Sbjct 152 VTEFFHTTVESVDFQKDSEKSRQEINFWVESQSQGKIKELFGKEAIDNSTVLVLVNAVYF 211
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYA 120
KA W F+ T F +N+ +E + VK M Q+ + G + + ++L M YA
Sbjct 212 KAKWEREFNSENTVDASFCLNE-----NEKKTVKMMNQKGKFRIGFIDELQAQILEMKYA 266
Query 121 DPRCRLYLFLP-------NSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIK 173
+ + + LP NS+ QEL++K E+L++ + S+ + +S P
Sbjct 267 MGKLSMLVLLPSCSEDNVNSL----QELEKKI-NHEKLLAWSSSENLSEKPVAISFPQFN 321
Query 174 LAAERN------QVGLVDVY 187
L + +G+ DV+
Sbjct 322 LEDSYDLKSILQDMGIKDVF 341
> dre:335229 serpinb1l4, fk92d08, wu:fk92d08, zgc:77645; serpin
peptidase inhibitor, clade B (ovalbumin), member 1, like 4;
K13963 serpin B
Length=393
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 51/163 (31%), Positives = 80/163 (49%), Gaps = 10/163 (6%)
Query 14 DFED-SAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF++ S A+ IN +VE T+ IK ++P+ A+ +R+VLVNA+YFK W F
Sbjct 138 DFKNKSEASRVNINKWVEKNTQEKIKDLLPSGAIDAMTRLVLVNAIYFKGNWEKKFPKEA 197
Query 73 TATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
T G F +N +T + VK M Q+ + F + ++L +PY + + LP+
Sbjct 198 TKDGQFKLNKNQT-----KPVKMMHQKAQFPFVVIPEINSQILELPYVGKNLSMLIILPD 252
Query 133 SIAAFEQELQ--QKPETTEELVSLVDNTPSSDVVLDLSLPLIK 173
I LQ +K T E+L+ +V +SLP K
Sbjct 253 EIEDATTGLQKLEKALTYEKLMQWTKVMRQQEV--QVSLPKFK 293
> hsa:5273 SERPINB10, PI10, bomapin; serpin peptidase inhibitor,
clade B (ovalbumin), member 10; K13963 serpin B
Length=397
Score = 73.2 bits (178), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 52/156 (33%), Positives = 83/156 (53%), Gaps = 19/156 (12%)
Query 25 INAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTATGPFYVNDTR 84
IN++VE +T+G I++++P ++ ++RM+LVNALYFK W F T PF +N+T
Sbjct 154 INSWVERQTEGKIQNLLPDDSVDSTTRMILVNALYFKGIWEHQFLVQNTTEKPFRINET- 212
Query 85 TGRSEVQQVKFMQQRLEEDFGLFESDKVKV--LSMPYADPRCRLYLFLPNSIAAFEQELQ 142
S+ Q+ FM+++L +F +K K L + Y L + LP I EQ
Sbjct 213 --TSKPVQMMFMKKKLH----IFHIEKPKAVGLQLYYKSRDLSLLILLPEDINGLEQ--L 264
Query 143 QKPETTEELVSLVDNTPSSDVV----LDLSLPLIKL 174
+K T E+L + S+D++ + L LP KL
Sbjct 265 EKAITYEKL----NEWTSADMMELYEVQLHLPKFKL 296
> hsa:89777 SERPINB12, MGC119247, MGC119248, YUKOPIN; serpin peptidase
inhibitor, clade B (ovalbumin), member 12; K13966 serpin
B11/12
Length=405
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 53/187 (28%), Positives = 92/187 (49%), Gaps = 18/187 (9%)
Query 12 STDFEDS-AAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
S DF+ + + IN +VE +++G IK + A+ + +VLVNA+YFKA W + F
Sbjct 144 SVDFQKNPEKSRQEINFWVECQSQGKIKELFSKDAINAETVLVLVNAVYFKAKWETYFDH 203
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
T PF +N +E + VK M Q+ G E K ++L M Y + +++ L
Sbjct 204 ENTVDAPFCLN-----ANENKSVKMMTQKGLYRIGFIEEVKAQILEMRYTKGKLSMFVLL 258
Query 131 P----NSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERN------Q 180
P +++ E EL++K T E++V+ + S+ + LS P L +
Sbjct 259 PSHSKDNLKGLE-ELERKI-TYEKMVAWSSSENMSEESVVLSFPRFTLEDSYDLNSILQD 316
Query 181 VGLVDVY 187
+G+ D++
Sbjct 317 MGITDIF 323
> mmu:67931 Serpini2, 1810006A24Rik, Spi14; serine (or cysteine)
peptidase inhibitor, clade I, member 2
Length=405
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 60/128 (46%), Gaps = 7/128 (5%)
Query 14 DFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRT 73
DF D+ +A AI+ +VE KT G IK++ P +R+VLVNA+YFK W F T
Sbjct 139 DFLDAKTSAQAISTWVESKTDGKIKNMFSEEEFGPLTRLVLVNAIYFKGDWKQKFRKEDT 198
Query 74 ATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKV--KVLSMPYADPRCRLYLFLP 131
F D T + V M+ L +G F + +VL +PY L + LP
Sbjct 199 EMTDFTKKDGSTVK-----VPMMKALLRAQYGYFSQSSMTCQVLELPYKADEFSLVIILP 253
Query 132 NSIAAFEQ 139
+ E+
Sbjct 254 TEDTSIEE 261
> dre:569051 im:7148243; K13963 serpin B
Length=440
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 51/164 (31%), Positives = 77/164 (46%), Gaps = 8/164 (4%)
Query 14 DFED-SAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF++ S AA IN +VE T+ IK ++P+ A+ +R+VLVNA+YFK W F
Sbjct 181 DFKNKSEAARVNINTWVEKNTQEKIKDLLPSGAIDAMTRLVLVNAIYFKGNWERKFPKEA 240
Query 73 TATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
T G F +N +T + VK M Q+ +VL +PY + + LP+
Sbjct 241 TNDGQFKLNKNQT-----KPVKMMYQKAHFPLASIPEMNSQVLELPYVGKNLSMLIILPD 295
Query 133 SI--AAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKL 174
I A E +K T E+L+ + +SLP K+
Sbjct 296 QIEDATTGLEKLEKALTYEKLMEWTKPEVMRQQEVQVSLPKFKM 339
> tgo:TGME49_017430 protease inhibitor, putative (EC:2.7.11.1)
Length=439
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 52/205 (25%), Positives = 100/205 (48%), Gaps = 23/205 (11%)
Query 6 LNCDAVSTDFE-DSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPW 64
L+ +A+ +F+ +S IN +V TK I ++P +A+TP + ++LV LYFK PW
Sbjct 128 LHTEALLANFKTNSNGEREKINEWVCSVTKRKIVDLLPPAAVTPETTLLLVGTLYFKGPW 187
Query 65 ASPFSPHRTAT-GPFYVNDTRTGRSEVQQVKFMQQRL----EEDFGLFESDK----VKVL 115
PF P ++ FY + ++FM+ +G +D+ + +L
Sbjct 188 LKPFVPCECSSLSKFYRQGPSGATISQEGIRFMESTQVCSGALRYGFKHTDRPGFGLTLL 247
Query 116 SMPYADPRCRLYLFLPNS---IAAFEQELQQKPETTEELV-SLVDNTPS--SDVVLDLSL 169
+PY D + + F+P+ +A E +++P+ +LV + D++ + DV L + L
Sbjct 248 EVPYIDIQSSMVFFMPDKPTDLAELEMMWREQPDLLNDLVQGMADSSGTELQDVELTIRL 307
Query 170 PLIKLAAER-------NQVGLVDVY 187
P +K++ + +G+ DV+
Sbjct 308 PYLKVSGDTISLTSALESLGVTDVF 332
> mmu:20723 Serpinb9, BB283241, CAP-3, CAP3, PI-9, PI9, Spi6,
ovalbumin; serine (or cysteine) peptidase inhibitor, clade B,
member 9; K13963 serpin B
Length=374
Score = 69.7 bits (169), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 91/184 (49%), Gaps = 23/184 (12%)
Query 16 EDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTAT 75
E++ + IN +V +T+G I ++ ++ +R+VL+NALYFK W PF+ T
Sbjct 123 EEAEVSRQHINTWVSKQTEGKIPELLSGGSVDSETRLVLINALYFKGKWHQPFNKEYTMD 182
Query 76 GPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPNS-- 133
PF +N + E + V+ M + + + + +VL MPY L + LP+
Sbjct 183 MPFKIN-----KDEKRPVQMMCREDTYNLAYVKEVQAQVLVMPYEGMELSLVVLLPDEGV 237
Query 134 -IAAFEQELQQKPETTEELVSLV--DNTPSSDVVLDLSLPLIKLAAERN------QVGLV 184
++ E L T E+L + + D S+DV ++ LP KL + + ++G+V
Sbjct 238 DLSKVENNL-----TFEKLTAWMEADFMKSTDV--EVFLPKFKLQEDYDMESLFQRLGVV 290
Query 185 DVYK 188
DV++
Sbjct 291 DVFQ 294
> hsa:5276 SERPINI2, MEPI, PANCPIN, PI14, TSA2004; serpin peptidase
inhibitor, clade I (pancpin), member 2
Length=405
Score = 69.3 bits (168), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 48/163 (29%), Positives = 77/163 (47%), Gaps = 11/163 (6%)
Query 14 DFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRT 73
DF+D+ A A I+ +VE KT G IK + P +R+VLVNA+YFK W F T
Sbjct 139 DFQDAKACAEMISTWVERKTDGKIKDMFSGEEFGPLTRLVLVNAIYFKGDWKQKFRKEDT 198
Query 74 ATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKV--KVLSMPYADPRCRLYLFLP 131
+N T+ S V ++ M+ L +G F + +VL + Y L + LP
Sbjct 199 Q----LINFTKKNGSTV-KIPMMKALLRTKYGYFSESSLNYQVLELSYKGDEFSLIIILP 253
Query 132 NSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKL 174
+ E +K T ++++ + +V ++SLP K+
Sbjct 254 AE--GMDIEEVEKLITAQQILKWLSEMQEEEV--EISLPRFKV 292
> xla:444669 serpini1, MGC84260, neuroserpin; serpin peptidase
inhibitor, clade I (neuroserpin), member 1
Length=410
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 62/125 (49%), Gaps = 11/125 (8%)
Query 12 STDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPH 71
+ DF +A A+ IN +VE T I+ + A +++VLVNALYFK W S F P
Sbjct 138 NVDFSQGSAVASHINLWVENHTNNRIRDLFTADDFNNLTKLVLVNALYFKGNWKSQFRPE 197
Query 72 RTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLF-----ESDKV-KVLSMPYADPRCR 125
T T F T+ SEV Q+ M Q+ E +G F E+ V +VL +PY
Sbjct 198 NTRTFSF----TKDDESEV-QIPMMYQKGEFYYGEFTDGSNEAGGVYQVLELPYEGEEIS 252
Query 126 LYLFL 130
L + L
Sbjct 253 LIIIL 257
> mmu:20706 Serpinb9b, 1600019A21Rik, R86, SPI-CI, Spi10, ovalbumin;
serine (or cysteine) peptidase inhibitor, clade B, member
9b; K13963 serpin B
Length=377
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 50/184 (27%), Positives = 84/184 (45%), Gaps = 19/184 (10%)
Query 15 FEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTA 74
F+ + + IN +V +T+G I ++ ++ +R+VLVNALYFK WA F T
Sbjct 122 FKAAVESRQCINTWVSKQTEGKIPELLADDSVNFQTRLVLVNALYFKGMWACQFCKESTR 181
Query 75 TGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPNS- 133
PFY+N + E + V+ M Q F + ++L MPY L + LP
Sbjct 182 EMPFYIN-----KDEKRPVQMMCQTDTFMFAFVDELPARLLIMPYEGMELSLMVLLPEKG 236
Query 134 --IAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAERNQ------VGLVD 185
++ E +L T E+L++ + + LP KL + +G+VD
Sbjct 237 VDLSKVENDL-----TFEKLIAWTKPDIMWSTEVKVFLPKFKLQEDYEMKSVLQCLGIVD 291
Query 186 VYKR 189
V+++
Sbjct 292 VFEK 295
> mmu:20717 Serpina3m, 3e46, AI195004, MMCM7, MMSPi2.4, Spi-2l,
Spi-2rs1, Spi2-rs1, Spi2.4, spi2; serine (or cysteine) peptidase
inhibitor, clade A, member 3M; K04525 serpin peptidase
inhibitor, clade A, member 3
Length=418
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 43/139 (30%), Positives = 68/139 (48%), Gaps = 7/139 (5%)
Query 9 DAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPF 68
+A + DF+ A IN +V +T+G IK ++ S L + MVLVN +YFK W F
Sbjct 162 EAFTADFQQPTEATKLINDYVSNQTQGMIKKLI--SELDDRTLMVLVNYIYFKGKWKISF 219
Query 69 SPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYL 128
P T FY+++ R+ + + ++KF+ R D E VL + Y L++
Sbjct 220 DPQDTFESEFYLDEKRSVKVPMMKMKFLTTRHFRD----EELSCSVLELKYTGNASALFI 275
Query 129 FLPNSIAAFEQELQQKPET 147
LP+ + E +PET
Sbjct 276 -LPDQGRMQQVEASLQPET 293
> mmu:18788 Serpinb2, PAI-2, Planh2, ovalbumin; serine (or cysteine)
peptidase inhibitor, clade B, member 2; K13963 serpin
B
Length=415
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 67/124 (54%), Gaps = 7/124 (5%)
Query 12 STDFEDSAAAA-AAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSP 70
+ DF + A A IN++V+ +TKG I +++P ++ ++MVLVNA+YFK W +PF
Sbjct 155 AVDFLECAEEAREKINSWVKTQTKGEIPNLLPEGSVDEDTKMVLVNAVYFKGKWKTPFEK 214
Query 71 HRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFL 130
PF VN + S Q+ F+ +L + G + K ++L +P+ + L L
Sbjct 215 KLNGLYPFRVN---SHESIPVQMMFLHAKL--NIGYIKDLKTQILELPHTG-NISMLLLL 268
Query 131 PNSI 134
P+ I
Sbjct 269 PDEI 272
> xla:379771 serpinc1, MGC52682; serpin peptidase inhibitor, clade
C (antithrombin), member 1; K03911 antithrombin III
Length=456
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/172 (28%), Positives = 82/172 (47%), Gaps = 21/172 (12%)
Query 25 INAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTATGPFYVNDTR 84
IN +V KT+ I ++P A+TP + +VL+NA+YFK W S F+ T F+
Sbjct 209 INNWVSNKTEKRITDVIPKDAITPDTVLVLINAIYFKGLWKSKFNSENTKMDQFH----- 263
Query 85 TGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPNS---IAAFEQEL 141
++ M Q +G F+ D V+VL +PY + L LP+ + EQ L
Sbjct 264 PAKNSNCLTATMYQEGTFRYGSFKDDGVQVLELPYKGDDITMVLVLPSQETPLTTVEQNL 323
Query 142 QQKPETTEELVSLVDNTPSSDVVLDLSLPLIK------LAAERNQVGLVDVY 187
T E+L + + S ++ L + LP + + + ++GLVD++
Sbjct 324 -----TLEKLGNWLQK--SRELQLSVYLPRFRVEDSFSVKEKLQEMGLVDLF 368
> mmu:380839 Serpinb1c, EIC, MGC129308, ovalbumin; serine (or
cysteine) peptidase inhibitor, clade B, member 1c; K13963 serpin
B
Length=375
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 52/194 (26%), Positives = 86/194 (44%), Gaps = 15/194 (7%)
Query 2 VSERLNCDAVSTDFEDSAA-AAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
+ + + D DF+ ++ A IN +V+G+T+ I+ + + +++VLVNA YF
Sbjct 108 IQKTYSADLALVDFQHASEDARKEINQWVKGQTEEKIQELFAVGVVDSMTKLVLVNATYF 167
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYA 120
K W F T PF R + + VK M + FG K KVL MPY
Sbjct 168 KGMWQKKFMARDTTDAPF-----RLSKKVTKTVKMMYLKNNLPFGYIPDLKCKVLEMPYQ 222
Query 121 DPRCRLYLFLPNSIAAFEQELQQ-KPETTEELVSLVDNTPSSDVVLDLSLPLIK------ 173
+ + LP I L++ + + T E + +N + DV + LP K
Sbjct 223 GGELSMVILLPEDIEDETTGLEEIEKQLTLEKLQECENLQNIDVC--VKLPKFKMEESYI 280
Query 174 LAAERNQVGLVDVY 187
L + Q+G+ D++
Sbjct 281 LNSNLGQLGVQDLF 294
> mmu:241196 Serpinb13, 5430417G24, HUR7, HURPIN, PI13, headpin;
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin),
member 13; K13963 serpin B
Length=389
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/142 (27%), Positives = 70/142 (49%), Gaps = 7/142 (4%)
Query 14 DFEDSA-AAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF ++A + IN++VE +T +K + P +L S+++VL+N +YFK W F
Sbjct 135 DFVNAADESRKKINSWVESQTNVKVKDLFPEGSLNSSTKLVLINTVYFKGLWDREFKKEH 194
Query 73 TATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
T F++N + + V+ M +F E + K++ +PY + +++ LPN
Sbjct 195 TKEEDFWLNKNLS-----KPVQMMALCSSFNFTFLEDLQAKIVGIPYKNNDISMFVLLPN 249
Query 133 SIAAFEQEL-QQKPETTEELVS 153
I E+ + + PE E S
Sbjct 250 DIDGLEKIMDKMSPEKLVEWTS 271
> mmu:238568 Serpinb6d, Gm11390, OTTMUSG00000000712, SPI3D; serine
(or cysteine) peptidase inhibitor, clade B, member 6d
Length=375
Score = 65.9 bits (159), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 45/162 (27%), Positives = 76/162 (46%), Gaps = 8/162 (4%)
Query 14 DFE-DSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHR 72
DF+ D+ + IN +V T IK ++ ++ ++R+VLVN YFK W PF+
Sbjct 124 DFKGDTEQSRQHINTWVTKNTDEKIKDLLSPGSVNSNTRLVLVNDFYFKGYWEKPFNKED 183
Query 73 TATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
T PF R ++ V+ V+ M Q+ E K+L +PYA + + + LP+
Sbjct 184 TREMPF-----RVSKNVVKPVQMMFQKSTFKITYIEEISTKILLLPYAGNKLNMIIMLPD 238
Query 133 SIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKL 174
E + +K T E+ V ++ +++ LP KL
Sbjct 239 E--HVELRMLEKKMTYEKFVEWTSLDKMNEEEVEVFLPRFKL 278
> mmu:20725 Serpinb8, CAP-2, CAP2, NK10, Spi8, ovalbumin; serine
(or cysteine) peptidase inhibitor, clade B, member 8; K13965
serpin B8
Length=245
Score = 65.9 bits (159), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 59/117 (50%), Gaps = 6/117 (5%)
Query 16 EDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTAT 75
+D+ IN +V KT+G I ++ + P +++VLVNA+YFK W + F T
Sbjct 123 KDTEGCRKHINDWVSEKTEGKISEVLSPGTVCPLTKLVLVNAMYFKGKWKAQFDRKYTRG 182
Query 76 GPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLPN 132
PF N E + V+ M + + G + ++VL++PYA+ + + LP+
Sbjct 183 MPFKTNQ------EKKTVQMMFKHAKFKMGHVDEVNMQVLALPYAEEELSMVILLPD 233
> hsa:6906 SERPINA7, FLJ17820, TBG; serpin peptidase inhibitor,
clade A (alpha-1 antiproteinase, antitrypsin), member 7
Length=415
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 68/123 (55%), Gaps = 10/123 (8%)
Query 9 DAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPF 68
+ STDF + +AA IN+ VE +TKG + ++ L P++ MVLVN ++FKA WA+PF
Sbjct 157 EVFSTDFSNISAAKQEINSHVEMQTKGKVVGLI--QDLKPNTIMVLVNYIHFKAQWANPF 214
Query 69 SPHRTA-TGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESD-KVKVLSMPYADPRCRL 126
P +T + F ++ T T QV M Q +E+ + L + + VL M Y+ L
Sbjct 215 DPSKTEDSSSFLIDKTTT-----VQVPMMHQ-MEQYYHLVDMELNCTVLQMDYSKNALAL 268
Query 127 YLF 129
++
Sbjct 269 FVL 271
> mmu:20714 Serpina3k, 1300001I07Rik, D12Rp54, MMCM2, MMSpi2,
RP54, Spi-2, Spi2, contrapsin; serine (or cysteine) peptidase
inhibitor, clade A, member 3K; K04525 serpin peptidase inhibitor,
clade A, member 3
Length=418
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 49/169 (28%), Positives = 79/169 (46%), Gaps = 11/169 (6%)
Query 9 DAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPF 68
+A + DF+ A IN +V +T+G IK ++ S L + MVLVN +YFK W F
Sbjct 163 EAFTADFQQPTEAKNLINDYVSNQTQGMIKKLI--SELDDGTLMVLVNYIYFKGKWKISF 220
Query 69 SPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYL 128
P T FY+++ R+ + + ++K + R D E VL + Y L L
Sbjct 221 DPQDTFESEFYLDEKRSVKVPMMKMKLLTARHFRD----EELSCSVLELKYTGNASAL-L 275
Query 129 FLPNSIAAFEQELQQKPETTEELVSLVDNTPSSDVVLDLSLPLIKLAAE 177
LP+ + E +PET + T S + +L+LP +A++
Sbjct 276 ILPDQGRMQQVEASLQPETLRKW----RKTLFSSQIEELNLPKFSIASD 320
> xla:495377 serpina3; serpin peptidase inhibitor, clade A (alpha-1
antiproteinase, antitrypsin), member 3
Length=303
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 70/133 (52%), Gaps = 12/133 (9%)
Query 1 QVSERLNCDAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
++ + + + F++ A A IN +V KT G I+ +V L+ ++++L+N + F
Sbjct 42 EIEHHYHAEIFPSHFKNPAEAEKQINDYVSNKTDGKIQELV--KDLSEETKLLLINYIVF 99
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESDKV--KVLSMP 118
KA W PFS H T +G F V+D T EVQ M + +E + F+ +K+ VL +P
Sbjct 100 KAEWEHPFSSHSTYSGKFSVDDNTT--VEVQ----MMYKTDE-YIFFKDEKIPCSVLQLP 152
Query 119 YADPRCRLYLFLP 131
Y + + L +P
Sbjct 153 YKN-NASMLLIVP 164
> hsa:8710 SERPINB7, DKFZp686D06190, MEGSIN, MGC120014, MGC120015;
serpin peptidase inhibitor, clade B (ovalbumin), member
7; K13964 serpin B7
Length=380
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 62/118 (52%), Gaps = 7/118 (5%)
Query 25 INAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPHRTATGPFYVNDTR 84
IN +VE +T G IK+++ ++ S+ MVLVNA+YFK W S F+ T F +
Sbjct 142 INKWVENETHGKIKNVIGEGGISSSAVMVLVNAVYFKGKWQSAFTKSETINCHF-----K 196
Query 85 TGRSEVQQVKFMQQRLEEDFGLFESDKVKVLSMPYADPRCRLYLFLP-NSIAAFEQEL 141
+ + + V M Q + + + E +K+L + Y + +Y+ LP N ++ E +L
Sbjct 197 SPKCSGKAVAMMHQERKFNLSVIEDPSMKILELRY-NGGINMYVLLPENDLSEIENKL 253
> dre:100334733 hypothetical protein LOC100334733
Length=383
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/139 (30%), Positives = 69/139 (49%), Gaps = 12/139 (8%)
Query 1 QVSERLNCDAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYF 60
Q+ N + + DF A + IN FV KT + ++ ++ P ++M+L+N +++
Sbjct 133 QIQRFFNAEVLRVDFSKPAVCRSLINEFVSRKTGRKVLEML--ESVEPLTQMMLLNTIFY 190
Query 61 KAPWASPFSPHRTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDFGLFESD--KVKVLSMP 118
K W PF+P+ T FYV+ + + QV M LEE F + E + +VL +P
Sbjct 191 KGDWERPFNPNNTEKSRFYVD-----KYNIVQVPMMM--LEEKFSVVEDRDLRARVLRLP 243
Query 119 YADPRCRLYLFLPNSIAAF 137
Y L L LPN+ A +
Sbjct 244 YRGGASMLIL-LPNADADY 261
> dre:557627 serpini1, serpini1l, si:ch211-167c22.4; serpin peptidase
inhibitor, clade I (neuroserpin), member 1
Length=415
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 44/128 (34%), Positives = 63/128 (49%), Gaps = 15/128 (11%)
Query 12 STDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPFSPH 71
+ DF S A A IN++V T+ I+++V A + S+ ++LVNA+YF+ W + F P
Sbjct 140 TVDFSQSTAVAERINSWVLNHTESKIQNLVSAEDFSSSTMIMLVNAVYFRGSWKNQFRPE 199
Query 72 RTATGPFYVNDTRTGRSEVQQVKFMQQRLEEDF-------GLFESDKV-KVLSMPYADPR 123
T T F TR SEVQ + QQ DF G E+ V +VL M Y
Sbjct 200 NTRTFSF----TRDDGSEVQTLMMYQQG---DFYYGEFSDGTTEAGGVYQVLEMLYEGED 252
Query 124 CRLYLFLP 131
+ + LP
Sbjct 253 MSMMIVLP 260
> hsa:12 SERPINA3, AACT, ACT, GIG25, MGC88254; serpin peptidase
inhibitor, clade A (alpha-1 antiproteinase, antitrypsin),
member 3; K04525 serpin peptidase inhibitor, clade A, member
3
Length=423
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/76 (40%), Positives = 45/76 (59%), Gaps = 2/76 (2%)
Query 9 DAVSTDFEDSAAAAAAINAFVEGKTKGHIKHIVPASALTPSSRMVLVNALYFKAPWASPF 68
+A +TDF+DSAAA IN +V+ T+G I ++ L + MVLVN ++FKA W PF
Sbjct 164 EAFATDFQDSAAAKKLINDYVKNGTRGKITDLI--KDLDSQTMMVLVNYIFFKAKWEMPF 221
Query 69 SPHRTATGPFYVNDTR 84
P T FY++ +
Sbjct 222 DPQDTHQSRFYLSKKK 237
Lambda K H
0.317 0.131 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5364689396
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40