bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1214_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
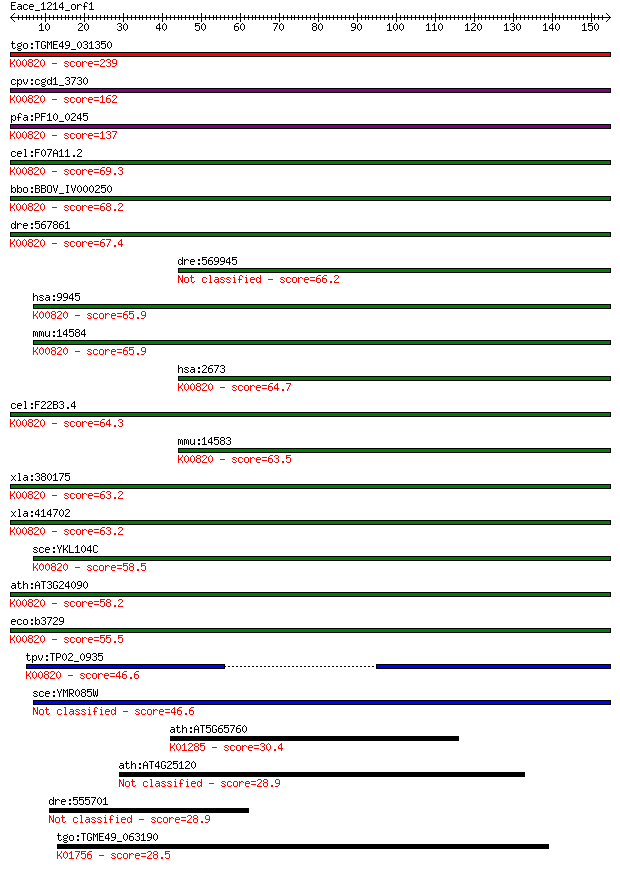
tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotrans... 239 3e-63
cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferas... 162 5e-40
pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferas... 137 2e-32
cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructos... 69.3 5e-12
bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate... 68.2 1e-11
dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase ... 67.4 2e-11
dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase ... 66.2 3e-11
hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphat... 65.9 5e-11
mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phospha... 65.9 6e-11
hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutam... 64.7 1e-10
cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose... 64.3 1e-10
mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,... 63.5 2e-10
xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosph... 63.2 3e-10
xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosph... 63.2 4e-10
sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fru... 58.5 9e-09
ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (iso... 58.2 1e-08
eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phos... 55.5 6e-08
tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferas... 46.6 3e-05
sce:YMR085W Putative protein of unknown function; YMR085W and ... 46.6 3e-05
ath:AT5G65760 serine carboxypeptidase S28 family protein; K012... 30.4 2.0
ath:AT4G25120 ATP binding / ATP-dependent DNA helicase/ DNA bi... 28.9 6.2
dre:555701 si:ch211-279l9.6 28.9 6.8
tgo:TGME49_063190 adenylosuccinate lyase, putative (EC:4.3.2.2... 28.5 8.4
> tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing), putative (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=603
Score = 239 bits (610), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 109/155 (70%), Positives = 137/155 (88%), Gaps = 1/155 (0%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLA 60
T+QYLML+DGEIA+VT++G+ QL +R +HRI+ ET+E SP+PF HWTLKEIFEQPQ+LA
Sbjct 196 TNQYLMLKDGEIAVVTAQGVGQLEATRPVHRIAKETIELSPEPFAHWTLKEIFEQPQALA 255
Query 61 RALNYGGRIMAHGNRVKLGGLDQNKD-LLAIRNLLLAGCGTSLYAGMYGEQLMQWLACFD 119
RA+NYGGRI + NRVKLGGLDQN++ LL +++LLL GCGTSLYAG+YGE LMQWL CFD
Sbjct 256 RAMNYGGRIAPYQNRVKLGGLDQNRESLLTVKSLLLCGCGTSLYAGIYGELLMQWLRCFD 315
Query 120 QVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
QVRA+DA+E++ + LP+ DAG+LLLSQSGETLDT+
Sbjct 316 QVRAVDASEVDIYHLPRQDAGVLLLSQSGETLDTV 350
> cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferase
(EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=696
Score = 162 bits (409), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 76/155 (49%), Positives = 112/155 (72%), Gaps = 1/155 (0%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLA 60
T+QY+ LQDGEIA+++ +GI++L+ + I E VE+SP P+ HWTLKEI++QP +LA
Sbjct 290 TNQYVALQDGEIALLSHEGINKLITPSRLLSIDHEKVESSPSPYLHWTLKEIYDQPHALA 349
Query 61 RALNYGGRIMAHGNRVKLGGLDQNKD-LLAIRNLLLAGCGTSLYAGMYGEQLMQWLACFD 119
R+LN+GGRI + N VKLGGLDQ D L ++N++L GCGTS +A ++ + LM+ ++ F+
Sbjct 350 RSLNFGGRISPYNNMVKLGGLDQRLDELKNVQNMILLGCGTSFHAALFAQLLMEHISGFN 409
Query 120 QVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
V A DA+E+ P+ AG + +SQSGET DT+
Sbjct 410 TVSAKDASEIFVTGFPREHAGAIAISQSGETADTV 444
> pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferase,
putative; K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=829
Score = 137 bits (344), Expect = 2e-32, Method: Composition-based stats.
Identities = 64/155 (41%), Positives = 99/155 (63%), Gaps = 1/155 (0%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLA 60
T++Y+ L++GEI ++ I+ L + + I ++ +P P+PHWT+KEI EQ +L+
Sbjct 421 TNEYISLKNGEILSISKDKINDLKLLKKVENIPEIAIQKTPHPYPHWTIKEIHEQSATLS 480
Query 61 RALNYGGRIMAHGNRVKLGGLDQN-KDLLAIRNLLLAGCGTSLYAGMYGEQLMQWLACFD 119
++LN GGR + + VKLGGLD +DL I NL+L GCGTS YA ++ + LM +L CF+
Sbjct 481 KSLNNGGRFSSGDHLVKLGGLDPYIQDLNKIENLVLVGCGTSYYAALFAKYLMNYLNCFN 540
Query 120 QVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
V+ +D + +PK G++ +SQSGET D I
Sbjct 541 TVQVMDPIDFNISVIPKEKEGVIFISQSGETRDVI 575
> cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=712
Score = 69.3 bits (168), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 49/164 (29%), Positives = 82/164 (50%), Gaps = 13/164 (7%)
Query 1 TSQYLMLQDGEIAIV---------TSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKE 51
T Q L L+D ++A V S+ D + R++ + E E F + KE
Sbjct 300 TKQVLFLEDDDVAFVEDGALTIHRISRHADNGEQKREVKLLEMELQEIMKGSFKTYMQKE 359
Query 52 IFEQPQSLARALNYGGRIMAHGNRVKLGGLDQN-KDLLAIRNLLLAGCGTSLYAGMYGEQ 110
IFEQP S+ + GR++ G +V LGG+ + D+ R L++ CGTS ++ + Q
Sbjct 360 IFEQPDSVVNTMR--GRVLPSG-QVVLGGIKEYLPDIKRCRRLIMVACGTSYHSAIACRQ 416
Query 111 LMQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+++ L+ V + + L++ + D L +SQSGET DT+
Sbjct 417 ILEELSELPVVVELASDFLDRETPIFRDDVCLFISQSGETADTL 460
> bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=723
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 53/196 (27%), Positives = 90/196 (45%), Gaps = 43/196 (21%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLA 60
T +++L+DG++ + +++L +R++ RI+ + + ++P+P W KE+F+QP
Sbjct 275 TDYFIVLEDGDVMELNYPVVEKLFTTREVCRITKQIIRSTPEPHQFWIQKEVFDQPLVAR 334
Query 61 RALNYGGRIMAHGNRVKL------GGLDQNKD-----------LLA------IR------ 91
AL + + A +V+L G +D D L++ +R
Sbjct 335 VALQHFKLVNA-SQKVRLLKQELEGLMDVTFDSEEHFIKEELELISSLNPDDVRFEIPVH 393
Query 92 ---NLL----------LAGCGTSLYAGMYGEQLMQWLACFDQVRAIDATELEQHSLPKHD 138
NLL L G+S A Y L Q A FD + D TEL+ + D
Sbjct 394 GDANLLKNIEMRHKINLVAAGSSHNAAKYVASLFQRSAMFDHIETDDPTELKMYRYTNPD 453
Query 139 AGILLLSQSGETLDTI 154
+ + +SQSGETLDT+
Sbjct 454 STFIYVSQSGETLDTV 469
> dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 48/163 (29%), Positives = 83/163 (50%), Gaps = 11/163 (6%)
Query 1 TSQYLMLQDGEIAIVTSK--GIDQLLES------RDMHRISAETVETSPDPFPHWTLKEI 52
T++ + L+D +IA V+ I ++ + R + + E + F + KEI
Sbjct 271 TNRVIFLEDDDIAAVSDGRLSIHRIKRTAGDHPARAIQTLQMELQQIMKGNFSSFMQKEI 330
Query 53 FEQPQSLARALNYGGRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSLYAGMYGEQL 111
FEQP+S+ + GR+ N V LGGL D K++ R L++ CGTS +AG+ Q+
Sbjct 331 FEQPESVVNTMR--GRVNFENNTVILGGLKDHIKEIQRCRRLIMIACGTSYHAGVATRQV 388
Query 112 MQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
++ L + + + L++++ D +SQSGET DT+
Sbjct 389 LEELTELPVMVELASDFLDRNTPVFRDDVCFFISQSGETADTL 431
> dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase
2 (EC:2.6.1.16)
Length=681
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 41/112 (36%), Positives = 62/112 (55%), Gaps = 3/112 (2%)
Query 44 FPHWTLKEIFEQPQSLARALNYGGRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSL 102
F + KEIFEQP+S+ + GRI N V LGGL D K++ R L++ GCGTS
Sbjct 321 FKAFMQKEIFEQPESVYNTMR--GRIFFDPNTVFLGGLTDHLKEIKRCRRLIMIGCGTSY 378
Query 103 YAGMYGEQLMQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+AG+ Q+++ L + + + L++ + D +SQSGET DT+
Sbjct 379 HAGVATRQILEELTELPVMVELASDFLDRITPVFRDDVCFFISQSGETADTL 430
> hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 46/149 (30%), Positives = 76/149 (51%), Gaps = 3/149 (2%)
Query 7 LQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLARALNYG 66
+ DG+++I K SR + + E + F + KEIFEQP+S+ +
Sbjct 285 VADGKLSIHRVKRSASDDPSRAIQTLQMELQQIMKGNFSAFMQKEIFEQPESVFNTMR-- 342
Query 67 GRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSLYAGMYGEQLMQWLACFDQVRAID 125
GR+ N V LGGL D K++ R L++ GCGTS +A + Q+++ L + +
Sbjct 343 GRVNFETNTVLLGGLKDHLKEIRRCRRLIVIGCGTSYHAAVATRQVLEELTELPVMVELA 402
Query 126 ATELEQHSLPKHDAGILLLSQSGETLDTI 154
+ L++++ D +SQSGET DT+
Sbjct 403 SDFLDRNTPVFRDDVCFFISQSGETADTL 431
> mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 46/149 (30%), Positives = 76/149 (51%), Gaps = 3/149 (2%)
Query 7 LQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLARALNYG 66
+ DG+++I K SR + + E + F + KEIFEQP+S+ +
Sbjct 285 VADGKLSIHRVKRSATDDPSRAIQTLQMELQQIMKGNFSAFMQKEIFEQPESVFNTMR-- 342
Query 67 GRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSLYAGMYGEQLMQWLACFDQVRAID 125
GR+ N V LGGL D K++ R L++ GCGTS +A + Q+++ L + +
Sbjct 343 GRVNFETNTVLLGGLKDHLKEIRRCRRLIVIGCGTSYHAAVATRQVLEELTELPVMVELA 402
Query 126 ATELEQHSLPKHDAGILLLSQSGETLDTI 154
+ L++++ D +SQSGET DT+
Sbjct 403 SDFLDRNTPVFRDDVCFFISQSGETADTL 431
> hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutamine--fructose-6-phosphate
transaminase 1 (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=681
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/112 (35%), Positives = 61/112 (54%), Gaps = 3/112 (2%)
Query 44 FPHWTLKEIFEQPQSLARALNYGGRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSL 102
F + KEIFEQP+S+ + GR+ V LGGL D K++ R L+L CGTS
Sbjct 321 FSSFMQKEIFEQPESVVNTMR--GRVNFDDYTVNLGGLKDHIKEIQRCRRLILIACGTSY 378
Query 103 YAGMYGEQLMQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+AG+ Q+++ L + + + L++++ D LSQSGET DT+
Sbjct 379 HAGVATRQVLEELTELPVMVELASDFLDRNTPVFRDDVCFFLSQSGETADTL 430
> cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=710
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 84/166 (50%), Gaps = 17/166 (10%)
Query 1 TSQYLMLQDGEIAIV---------TSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKE 51
T Q L L+D ++A V S+ D + R++ + E E + + KE
Sbjct 298 TKQVLFLEDDDVAFVEDGSLTIHRISRHADSGEQKREVQLLELEIQEIMKGSYKTFMQKE 357
Query 52 IFEQPQSLARALNYGGRIMAHGNRVKLGGLDQNKDLLAI---RNLLLAGCGTSLYAGMYG 108
IFEQP+S+ + GR++ G +V LGGL + L AI R L++ CGTS ++ +
Sbjct 358 IFEQPESVVNTMR--GRVLPSG-QVVLGGLKEY--LPAIKRCRRLIMVACGTSYHSAIAC 412
Query 109 EQLMQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
Q+++ L+ V + + L++ + + L +SQSGET DT+
Sbjct 413 RQVLEELSELPVVVELASDFLDRETPIFRNDVCLFISQSGETADTL 458
> mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,
GFAT1, GFAT1m, Gfpt; glutamine fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 61/112 (54%), Gaps = 3/112 (2%)
Query 44 FPHWTLKEIFEQPQSLARALNYGGRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSL 102
F + KEIFEQP+S+ + GR+ V LGGL D K++ R L+L CGTS
Sbjct 321 FSSFMQKEIFEQPESVVNTMR--GRVNFDDYTVNLGGLKDHIKEIQRCRRLILIACGTSY 378
Query 103 YAGMYGEQLMQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+AG+ Q+++ L + + + L++++ D +SQSGET DT+
Sbjct 379 HAGVATRQVLEELTELPVMVELASDFLDRNTPVFRDDVCFFISQSGETADTL 430
> xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/163 (28%), Positives = 79/163 (48%), Gaps = 11/163 (6%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVET--------SPDPFPHWTLKEI 52
T++ + L+D ++A V + R++ A ++T F + KEI
Sbjct 270 TNRVIFLEDDDVAAVVDGRLSIHRIKRNVGDHPARAIQTLQMELQQIMKGNFSSFMQKEI 329
Query 53 FEQPQSLARALNYGGRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSLYAGMYGEQL 111
FEQP+S+ + GR+ V LGGL D K++ R L+L CGTS +AG+ Q+
Sbjct 330 FEQPESVVNTMR--GRVNFDDLTVNLGGLKDHIKEIQRCRRLILIACGTSYHAGVATRQI 387
Query 112 MQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
++ L + + + L++ + D +SQSGET DT+
Sbjct 388 LEELTELPVMVELASDFLDRSTPVFRDDVCFFISQSGETADTL 430
> xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosphate
transaminase 1; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 47/163 (28%), Positives = 80/163 (49%), Gaps = 11/163 (6%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVET--------SPDPFPHWTLKEI 52
T++ + L+D ++A V + R++ A ++T F + KEI
Sbjct 270 TNRVIFLEDDDVAAVVDGRLSIHRIKRNVGDHPARAIQTLQMELQQIMKGNFSSFMQKEI 329
Query 53 FEQPQSLARALNYGGRIMAHGNRVKLGGL-DQNKDLLAIRNLLLAGCGTSLYAGMYGEQL 111
FEQP+S+ + GR+ V LGGL D K++ R L+L CGTS +AG+ Q+
Sbjct 330 FEQPESVVNTMR--GRLNFDDLTVNLGGLKDHIKEIQRCRRLILIACGTSYHAGVATRQI 387
Query 112 MQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
++ L + + + L++++ D +SQSGET DT+
Sbjct 388 LEELTELPVMVELASDFLDRNTPVFRDDVCFFISQSGETADTL 430
> sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=717
Score = 58.5 bits (140), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 73/152 (48%), Gaps = 9/152 (5%)
Query 7 LQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLARALNYG 66
+ DGE+ I S+ +R + + E + P+ H+ KEI+EQP+S +
Sbjct 315 IYDGELHIHRSRREVGASMTRSIQTLEMELAQIMKGPYDHFMQKEIYEQPESTFNTMR-- 372
Query 67 GRIMAHGNRVKLGGLDQNKDLLAI----RNLLLAGCGTSLYAGMYGEQLMQWLACFDQVR 122
GRI N+V LGGL K L + R L++ CGTS ++ + + + L+
Sbjct 373 GRIDYENNKVILGGL---KAWLPVVRRARRLIMIACGTSYHSCLATRAIFEELSDIPVSV 429
Query 123 AIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+ + L++ D + +SQSGET DT+
Sbjct 430 ELASDFLDRKCPVFRDDVCVFVSQSGETADTM 461
> ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (isomerizing)/
sugar binding / transaminase (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=695
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 52/177 (29%), Positives = 85/177 (48%), Gaps = 27/177 (15%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQL-LES---------------RDMHRISAETVETSPDPF 44
T + L+++DGE+ + G+ L E+ R + + E + S +
Sbjct 270 TKKVLVIEDGEVVNLKDGGVSILKFENERGRCNGLSRPASVERALSVLEMEVEQISKGKY 329
Query 45 PHWTLKEIFEQPQSLARALNYGGRIMAHGNR----VKLGGL-DQNKDLLAIRNLLLAGCG 99
H+ KEI EQP+SL + GR++ G+R V LGGL D K + R ++ GCG
Sbjct 330 DHYMQKEIHEQPESLTTTMR--GRLIRGGSRKTKTVLLGGLKDHLKTIRRSRRIVFIGCG 387
Query 100 TSLYAGMYGEQLMQWLACFDQVRAIDATEL--EQHSLPKHDAGILLLSQSGETLDTI 154
TS A + +++ L+ I A++L Q + + D + +SQSGET DT+
Sbjct 388 TSYNAALASRPILEELSGIPVSMEI-ASDLWDRQGPIYREDTAV-FVSQSGETADTL 442
> eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=609
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 45/163 (27%), Positives = 83/163 (50%), Gaps = 15/163 (9%)
Query 1 TSQYLMLQDGEIAIVTSKGIDQLLESRDMHRISAETVETS-------PDPFPHWTLKEIF 53
T +++ L++G+IA +T + ++ + + + + +E++ + H+ KEI+
Sbjct 201 TRRFIFLEEGDIAEITRRSVN--IFDKTGAEVKRQDIESNLQYDAGDKGIYRHYMQKEIY 258
Query 54 EQPQSLARALNYGGRIMAHGNRVKLGGLDQNKD--LLAIRNLLLAGCGTSLYAGMYGEQL 111
EQP ++ L GRI +HG +V L L N D L + ++ + CGTS +GM
Sbjct 259 EQPNAIKNTLT--GRI-SHG-QVDLSELGPNADELLSKVEHIQILACGTSYNSGMVSRYW 314
Query 112 MQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+ LA I + + S + ++ ++ LSQSGET DT+
Sbjct 315 FESLAGIPCDVEIASEFRYRKSAVRRNSLMITLSQSGETADTL 357
> tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferase;
K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=810
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 35/60 (58%), Gaps = 0/60 (0%)
Query 95 LAGCGTSLYAGMYGEQLMQWLACFDQVRAIDATELEQHSLPKHDAGILLLSQSGETLDTI 154
CG+SL+A Y +++Q + FD V A DA++L + D ++ +S SGETLD I
Sbjct 441 FVACGSSLHAATYVAKILQKIHHFDLVEADDASDLTLYRYHDKDVTVVHISNSGETLDCI 500
Score = 38.1 bits (87), Expect = 0.012, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 5 LMLQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQ 55
++L DG+I ++ + ++ ++ ++ +E +E S +P P+W KEI +Q
Sbjct 287 IVLNDGDILELSLENVESYYSQYNLLKLESEVIEESHEPHPNWYTKEILDQ 337
> sce:YMR085W Putative protein of unknown function; YMR085W and
adjacent ORF YMR084W are merged in related strains
Length=432
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/151 (25%), Positives = 73/151 (48%), Gaps = 7/151 (4%)
Query 7 LQDGEIAIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLARALNYG 66
+ DGE+ I SK + R + ++ E + P+ ++ KEI+EQ ++ +
Sbjct 30 IYDGELHIHCSKIGSEDFSFRTVQKLELELSKIKKGPYDNFMQKEIYEQCETTKNVMR-- 87
Query 67 GRIMAHGNRVKLGGLDQ-NKDLLAIRNLLLAGCGTSLYAGMYGEQLMQWL--ACFDQVRA 123
GR+ A NRV LGGL+ +L + +++ S ++ + + + L + A
Sbjct 88 GRVDAFTNRVVLGGLENWLTELRRAKRIIMIASKASFHSCLAARPIFEELMEVPVNVELA 147
Query 124 IDATELEQHSLPKHDAGILLLSQSGETLDTI 154
+D + + ++D I +S+SGET DTI
Sbjct 148 LDFVD-RNCCIFRNDVCI-FVSRSGETTDTI 176
> ath:AT5G65760 serine carboxypeptidase S28 family protein; K01285
lysosomal Pro-X carboxypeptidase [EC:3.4.16.2]
Length=515
Score = 30.4 bits (67), Expect = 2.0, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 33/82 (40%), Gaps = 8/82 (9%)
Query 42 DPFPHWTLKEIFEQPQSLARALNYGGRIMAHGNRVKLGGLDQNKDL--LAI------RNL 93
D P + +F + + ++ YG R A+ N L L + L A+ RNL
Sbjct 120 DIAPKFGALLVFPEHRYYGESMPYGSREEAYKNATTLSYLTTEQALADFAVFVTDLKRNL 179
Query 94 LLAGCGTSLYAGMYGEQLMQWL 115
C L+ G YG L W+
Sbjct 180 SAEACPVVLFGGSYGGMLAAWM 201
> ath:AT4G25120 ATP binding / ATP-dependent DNA helicase/ DNA
binding / hydrolase
Length=1149
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 26/107 (24%), Positives = 46/107 (42%), Gaps = 16/107 (14%)
Query 29 MHRISAETVETSPDPFPHWTLKEIFEQPQSLARALNY--GGRIMAHGNRVKLGGLDQNKD 86
+ R E F +W K+ F++P+ L + + G R+ N+ +KD
Sbjct 1008 LKRFDVEVRSVVSHLFHNWAKKQAFQEPKRLIDKVRFVIGERLAIKKNK--------HKD 1059
Query 87 LL-AIRNLLLAGCGTSLYAGMYGEQLMQWLACFDQVRAIDATELEQH 132
+L A+++ L TS A Y E +++W RA E ++H
Sbjct 1060 VLRALKSSL-----TSEEAFQYAEHVLRWEQLPADTRAHIVREKQEH 1101
> dre:555701 si:ch211-279l9.6
Length=1673
Score = 28.9 bits (63), Expect = 6.8, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Query 11 EIAIVTSKGIDQLLESRDMHRI-SAETVETSPDPFPHW--TLKEIFEQPQSLAR 61
E+++V++ +D L + H+ SA V + PD FPH + K+ F +L+R
Sbjct 1351 EMSLVSNLVVDVQLSGEEQHQSKSAPEVVSPPDEFPHTCASCKKTFRHAATLSR 1404
> tgo:TGME49_063190 adenylosuccinate lyase, putative (EC:4.3.2.2);
K01756 adenylosuccinate lyase [EC:4.3.2.2]
Length=500
Score = 28.5 bits (62), Expect = 8.4, Method: Composition-based stats.
Identities = 38/138 (27%), Positives = 53/138 (38%), Gaps = 27/138 (19%)
Query 13 AIVTSKGIDQLLESRDMHRISAETVETSPDPFPHWTLKEIFEQPQSLARALNYGGRI-MA 71
A V S+ + L + + R SA SP PFP + E+ ++ R + GR M
Sbjct 53 AFVMSEWLACLATNARVQRQSAADASESPAPFP-----SLSEEGKAALRQM---GRFSMD 104
Query 72 HGNRVKLGGLDQNKDLLAIRNLLLAGCGTS---------LYAGMYGEQL--MQWLACFDQ 120
RVK N DL A+ +L TS + G E + + W CF +
Sbjct 105 DVRRVKEVEKTANHDLKAVEYVLKGKLETSEATMPLAALTHFGCTSEDVNNLSWALCFKE 164
Query 121 VRAIDATELEQHSLPKHD 138
EQH LP D
Sbjct 165 AT-------EQHLLPAVD 175
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40