bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1234_orf2
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
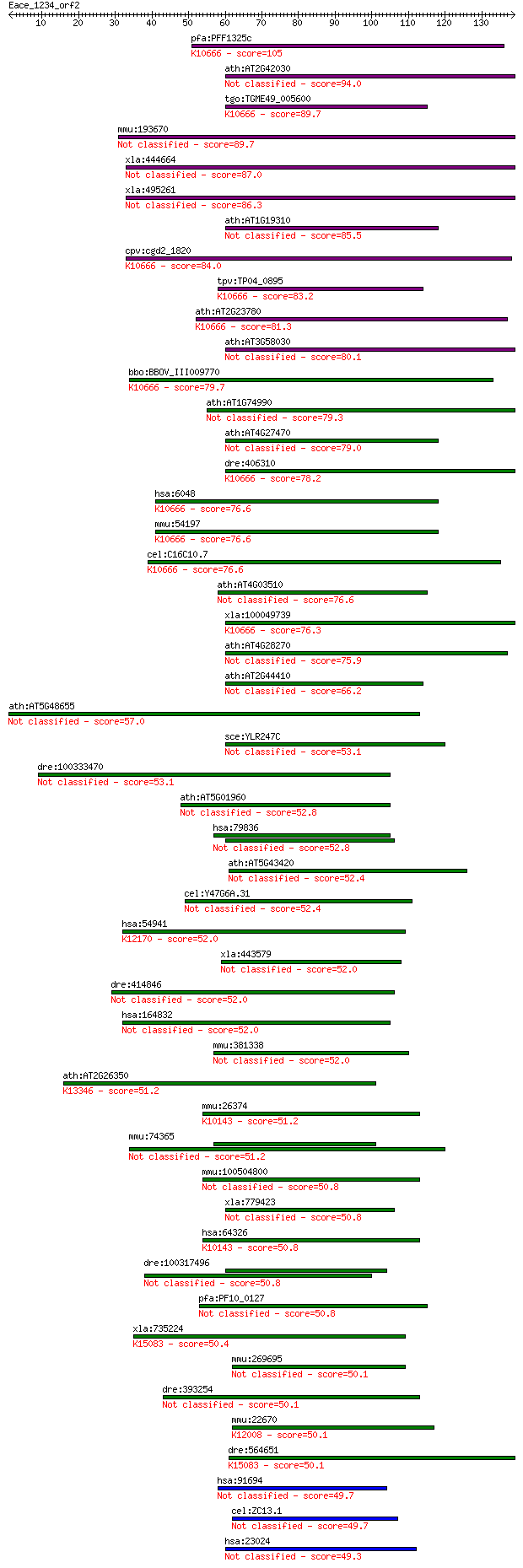
pfa:PFF1325c c3h4-type ring finger protein, putative; K10666 E... 105 4e-23
ath:AT2G42030 zinc finger (C3HC4-type RING finger) family protein 94.0 1e-19
tgo:TGME49_005600 zinc finger (C3HC4 RING finger) protein, put... 89.7 2e-18
mmu:193670 Rnf185, 1700022N24Rik, AL033296, MGC19394; ring fin... 89.7 2e-18
xla:444664 MGC84239 protein 87.0 1e-17
xla:495261 rnf185, xrnf185; ring finger protein 185 86.3
ath:AT1G19310 zinc finger (C3HC4-type RING finger) family protein 85.5 4e-17
cpv:cgd2_1820 hypothetical protein ; K10666 E3 ubiquitin-prote... 84.0 1e-16
tpv:TP04_0895 hypothetical protein; K10666 E3 ubiquitin-protei... 83.2 2e-16
ath:AT2G23780 zinc finger (C3HC4-type RING finger) family prot... 81.3 9e-16
ath:AT3G58030 zinc finger (C3HC4-type RING finger) family protein 80.1 2e-15
bbo:BBOV_III009770 17.m07848; hypothetical protein; K10666 E3 ... 79.7 2e-15
ath:AT1G74990 zinc finger (C3HC4-type RING finger) family protein 79.3 3e-15
ath:AT4G27470 zinc finger (C3HC4-type RING finger) family protein 79.0 4e-15
dre:406310 rnf185; zgc:73070; K10666 E3 ubiquitin-protein liga... 78.2 8e-15
hsa:6048 RNF5, RING5, RMA1; ring finger protein 5; K10666 E3 u... 76.6 2e-14
mmu:54197 Rnf5, 2410131O05Rik, AA407576, NG2; ring finger prot... 76.6 2e-14
cel:C16C10.7 rnf-5; RiNg Finger protein family member (rnf-5);... 76.6 2e-14
ath:AT4G03510 RMA1; RMA1; protein binding / ubiquitin-protein ... 76.6 2e-14
xla:100049739 rnf5; ring finger protein 5; K10666 E3 ubiquitin... 76.3 3e-14
ath:AT4G28270 zinc finger (C3HC4-type RING finger) family protein 75.9 4e-14
ath:AT2G44410 protein binding / zinc ion binding 66.2 3e-11
ath:AT5G48655 zinc finger (C3HC4-type RING finger) family protein 57.0 2e-08
sce:YLR247C IRC20; Irc20p (EC:3.6.1.-) 53.1 2e-07
dre:100333470 ReO_6-like 53.1 2e-07
ath:AT5G01960 zinc finger (C3HC4-type RING finger) family protein 52.8 4e-07
hsa:79836 LONRF3, FLJ22612, MGC119463, MGC119465, RNF127; LON ... 52.8 4e-07
ath:AT5G43420 zinc finger (C3HC4-type RING finger) family protein 52.4 4e-07
cel:Y47G6A.31 hypothetical protein 52.4 4e-07
hsa:54941 RNF125, FLJ20456, MGC21737, TRAC1; ring finger prote... 52.0 5e-07
xla:443579 rnf219; ring finger protein 219 52.0
dre:414846 rnf141, zgc:86917; ring finger protein 141 52.0
hsa:164832 LONRF2, FLJ45273, MGC126711, MGC126713, RNF192; LON... 52.0 6e-07
mmu:381338 Lonrf2, 2900060P06Rik, AI851349; LON peptidase N-te... 52.0 6e-07
ath:AT2G26350 PEX10; PEX10; protein binding / zinc ion binding... 51.2 9e-07
mmu:26374 Rfwd2, AI316802, C80879, Cop1; ring finger and WD re... 51.2 1e-06
mmu:74365 Lonrf3, 4932412G04Rik, 5730439E01Rik, A830039N02Rik,... 51.2 1e-06
mmu:100504800 e3 ubiquitin-protein ligase RFWD2-like 50.8 1e-06
xla:779423 pdzrn3, MGC80196; PDZ domain containing ring finger 3 50.8
hsa:64326 RFWD2, COP1, FLJ10416, RNF200; ring finger and WD re... 50.8 1e-06
dre:100317496 si:dkey-258f14.5 50.8 1e-06
pfa:PF10_0127 conserved Plasmodium protein 50.8 1e-06
xla:735224 hltf, MGC131155, smarca3; helicase-like transcripti... 50.4 2e-06
mmu:269695 Rnft2, AW049082, B830028P19Rik, Tmem118; ring finge... 50.1 2e-06
dre:393254 MGC56368, wu:fc04b02; zgc:56368 50.1 2e-06
mmu:22670 Trim26, AI462198, Zfp173, Zfp1736; tripartite motif-... 50.1 2e-06
dre:564651 hltf; helicase-like transcription factor-like; K150... 50.1 2e-06
hsa:91694 LONRF1, FLJ23749, RNF191; LON peptidase N-terminal d... 49.7 2e-06
cel:ZC13.1 hypothetical protein 49.7 2e-06
hsa:23024 PDZRN3, LNX3, SEMACAP3; PDZ domain containing ring f... 49.3 3e-06
> pfa:PFF1325c c3h4-type ring finger protein, putative; K10666
E3 ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=449
Score = 105 bits (262), Expect = 4e-23, Method: Composition-based stats.
Identities = 40/85 (47%), Positives = 54/85 (63%), Gaps = 0/85 (0%)
Query 51 ATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTP 110
AT E + FEC IC D++ DPVVT+CGHLFCW CL W++++ DCP+CKA VS + V P
Sbjct 279 ATENESRNTFECNICFDDVRDPVVTKCGHLFCWLCLSAWIKKNNDCPVCKAEVSKENVIP 338
Query 111 IYGRSSSPKSHAHTRHFSSETPPSR 135
+YGR + H + + P R
Sbjct 339 LYGRGKNSSDHKYAQPEEPRPTPKR 363
> ath:AT2G42030 zinc finger (C3HC4-type RING finger) family protein
Length=425
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 53/81 (65%), Gaps = 5/81 (6%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ--QSGDCPICKAAVSADTVTPIYGRSSS 117
F+C IC+D DPVVT CGHL+CW CL+ WLQ ++ +CP+CK VS TVTPIYGR
Sbjct 139 FDCYICLDLSKDPVVTNCGHLYCWSCLYQWLQVSEAKECPVCKGEVSVKTVTPIYGRGIQ 198
Query 118 PKSHAHTRHFSSETPPSRPRA 138
+ + S+ PSRP+A
Sbjct 199 KR---ESEEVSNTKIPSRPQA 216
> tgo:TGME49_005600 zinc finger (C3HC4 RING finger) protein, putative
; K10666 E3 ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=484
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 42/56 (75%), Gaps = 1/56 (1%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQ-SGDCPICKAAVSADTVTPIYGR 114
FEC IC DE DPVVTRCGHLFCW CLH WL++ + +CP+CKA + V PIYGR
Sbjct 336 FECNICFDEATDPVVTRCGHLFCWTCLHAWLRRGTYECPVCKAHTTVRNVIPIYGR 391
> mmu:193670 Rnf185, 1700022N24Rik, AL033296, MGC19394; ring finger
protein 185
Length=228
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 49/112 (43%), Positives = 66/112 (58%), Gaps = 10/112 (8%)
Query 31 TSSSSSSNSNSSSSSSSSRGATRKEGA-SPFECCICMDEIHDPVVTRCGHLFCWGCLHLW 89
++S+S+ NSN+ S SS G G S FEC IC+D D V++ CGHLFCW CLH W
Sbjct 43 SASASTENSNAGGPSGSSNGTGESGGQDSTFECNICLDTAKDAVISLCGHLFCWPCLHQW 102
Query 90 LQQSGD---CPICKAAVSADTVTPIYGRSSSPKSHAHTRHFSSETPPSRPRA 138
L+ + CP+CKA +S D V P+YGR S+ + + TPP RP+
Sbjct 103 LETRPNRQVCPVCKAGISRDKVIPLYGRGSTGQQDPREK-----TPP-RPQG 148
> xla:444664 MGC84239 protein
Length=189
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 45/109 (41%), Positives = 65/109 (59%), Gaps = 10/109 (9%)
Query 33 SSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQ 92
S+S+ NS +S S+ G T + ++ FEC IC+D D V++ CGHLFCW CLH WL+
Sbjct 9 SASAENSGPGGASGSTNGETSSQDST-FECNICLDNAKDAVISLCGHLFCWPCLHQWLET 67
Query 93 SGD---CPICKAAVSADTVTPIYGRSSSPKSHAHTRHFSSETPPSRPRA 138
+ CP+CKA +S + V P+YGR S+ + + TPP RP+
Sbjct 68 RPNRQVCPVCKAGISREKVIPLYGRGSTGQEDPREK-----TPP-RPQG 110
> xla:495261 rnf185, xrnf185; ring finger protein 185
Length=189
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 45/109 (41%), Positives = 65/109 (59%), Gaps = 10/109 (9%)
Query 33 SSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQ 92
S+S+ NS +S S+ G T + ++ FEC IC+D D V++ CGHLFCW CLH WL+
Sbjct 9 SASAENSGPGGASGSTNGETSSQDST-FECNICLDTAKDAVISLCGHLFCWPCLHQWLET 67
Query 93 SGD---CPICKAAVSADTVTPIYGRSSSPKSHAHTRHFSSETPPSRPRA 138
+ CP+CKA +S + V P+YGR S+ + + TPP RP+
Sbjct 68 RPNRQVCPVCKAGISREKVIPLYGRGSTGQEDPREK-----TPP-RPQG 110
> ath:AT1G19310 zinc finger (C3HC4-type RING finger) family protein
Length=226
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 35/61 (57%), Positives = 42/61 (68%), Gaps = 3/61 (4%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ---QSGDCPICKAAVSADTVTPIYGRSS 116
FEC IC+D DP+VT CGHLFCW CL+ WL QS DCP+CKA + D + P+YGR
Sbjct 21 FECNICLDLAQDPIVTLCGHLFCWPCLYKWLHLHSQSKDCPVCKAVIEEDRLVPLYGRGK 80
Query 117 S 117
S
Sbjct 81 S 81
> cpv:cgd2_1820 hypothetical protein ; K10666 E3 ubiquitin-protein
ligase RNF5 [EC:6.3.2.19]
Length=200
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/109 (36%), Positives = 60/109 (55%), Gaps = 5/109 (4%)
Query 33 SSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQ 92
+ S N S R + + + FEC IC + ++P+VTRCGHL+CW C+ WL +
Sbjct 13 DAKGSGLNGEQSDHHHRSEEKSKNYTSFECNICFENAYEPIVTRCGHLYCWSCICSWLDR 72
Query 93 S-GDCPICKAAVSADTVTPIYGRSSS---PKSHAHTRHFSSETPPSRPR 137
DCP+CKA V+++ V P+YGR + P+ R +E P +R R
Sbjct 73 GYEDCPVCKAGVNSENVIPLYGRGNENVDPRKKTKPRP-KAERPEARQR 120
> tpv:TP04_0895 hypothetical protein; K10666 E3 ubiquitin-protein
ligase RNF5 [EC:6.3.2.19]
Length=284
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 33/57 (57%), Positives = 43/57 (75%), Gaps = 1/57 (1%)
Query 58 SPFECCICMDEIHDPVVTRCGHLFCWGCLHLWL-QQSGDCPICKAAVSADTVTPIYG 113
S FEC IC DE+ DPVVTRCGHLFCW CL W+ +++ CPIC++ +S + V P+YG
Sbjct 19 SKFECNICFDEVKDPVVTRCGHLFCWSCLLSWMNRRNYQCPICQSGISRENVIPLYG 75
> ath:AT2G23780 zinc finger (C3HC4-type RING finger) family protein;
K10666 E3 ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=227
Score = 81.3 bits (199), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 37/88 (42%), Positives = 52/88 (59%), Gaps = 5/88 (5%)
Query 52 TRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ---QSGDCPICKAAVSADTV 108
T +G FEC IC + DP+VT CGHLFCW CL+ WL S +CP+CKA V D +
Sbjct 19 TNDQGGD-FECNICFELAQDPIVTLCGHLFCWPCLYRWLHHHSHSQECPVCKAVVQDDKL 77
Query 109 TPIYGRSSSPKSHAHTRHFSSETPPSRP 136
P+YGR + ++ ++ + P+RP
Sbjct 78 VPLYGRGKN-QTDPRSKRYPGLRIPNRP 104
> ath:AT3G58030 zinc finger (C3HC4-type RING finger) family protein
Length=436
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 39/82 (47%), Positives = 52/82 (63%), Gaps = 7/82 (8%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ--QSGDCPICKAAVSADTVTPIYGRSSS 117
F+C IC+D +PV+T CGHL+CW CL+ WLQ + +CP+CK V++ TVTPIYGR
Sbjct 137 FDCNICLDLSKEPVLTCCGHLYCWPCLYQWLQISDAKECPVCKGEVTSKTVTPIYGRG-- 194
Query 118 PKSHAHTRHFSSETP-PSRPRA 138
+H S +T P RP A
Sbjct 195 --NHKREIEESLDTKVPMRPHA 214
> bbo:BBOV_III009770 17.m07848; hypothetical protein; K10666 E3
ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=159
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/103 (35%), Positives = 59/103 (57%), Gaps = 11/103 (10%)
Query 34 SSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQS 93
S ++N ++ S + T ++C IC +++ DPVVTRCGHLFCW CL W+ +
Sbjct 9 SPPEDTNQTNKSPGDKKQT-------YDCNICFEDVVDPVVTRCGHLFCWQCLLTWINKP 61
Query 94 GD-CPICKAAVSADTVTPIYGR---SSSPKSHAHTRHFSSETP 132
D CP+C A ++ + V P+YGR ++ P++ S+E P
Sbjct 62 NDHCPVCHAGITKENVIPLYGRGQETNDPRNKPSEPRPSAERP 104
> ath:AT1G74990 zinc finger (C3HC4-type RING finger) family protein
Length=137
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/87 (43%), Positives = 53/87 (60%), Gaps = 4/87 (4%)
Query 55 EGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ---QSGDCPICKAAVSADTVTPI 111
+ ++ F C IC++ +P+VT CGHLFCW CL+ WL +S CP+CKA V DT+ P+
Sbjct 12 DASNNFGCNICLELAREPIVTLCGHLFCWPCLYKWLHYHSKSNHCPVCKALVKEDTLVPL 71
Query 112 YGRSSSPKSHAHTRHFSSETPPSRPRA 138
YG P S ++ S T P+RP A
Sbjct 72 YGM-GKPSSDPRSKLNSGVTVPNRPAA 97
> ath:AT4G27470 zinc finger (C3HC4-type RING finger) family protein
Length=243
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 43/68 (63%), Gaps = 10/68 (14%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWL----------QQSGDCPICKAAVSADTVT 109
F+C IC+D HDPVVT CGHLFCW C++ WL Q +CP+CK+ ++ ++
Sbjct 42 FDCNICLDTAHDPVVTLCGHLFCWPCIYKWLHVQLSSVSVDQHQNNCPVCKSNITITSLV 101
Query 110 PIYGRSSS 117
P+YGR S
Sbjct 102 PLYGRGMS 109
> dre:406310 rnf185; zgc:73070; K10666 E3 ubiquitin-protein ligase
RNF5 [EC:6.3.2.19]
Length=194
Score = 78.2 bits (191), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 37/82 (45%), Positives = 48/82 (58%), Gaps = 9/82 (10%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGD---CPICKAAVSADTVTPIYGRSS 116
FEC IC+D D V++ CGHLFCW CLH WL+ + CP+CKA +S D V P+YGR S
Sbjct 39 FECNICLDTSKDAVISLCGHLFCWPCLHQWLETRPNRQVCPVCKAGISRDKVIPLYGRGS 98
Query 117 SPKSHAHTRHFSSETPPSRPRA 138
+ + E P RP+
Sbjct 99 TGQQDPR------EKTPPRPQG 114
> hsa:6048 RNF5, RING5, RMA1; ring finger protein 5; K10666 E3
ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=180
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/82 (40%), Positives = 48/82 (58%), Gaps = 5/82 (6%)
Query 41 SSSSSSSSRGATRKEGAS--PFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ---QSGD 95
+ G R+ G + FEC IC++ + VV+ CGHL+CW CLH WL+ + +
Sbjct 4 AEEEDGGPEGPNRERGGAGATFECNICLETAREAVVSVCGHLYCWPCLHQWLETRPERQE 63
Query 96 CPICKAAVSADTVTPIYGRSSS 117
CP+CKA +S + V P+YGR S
Sbjct 64 CPVCKAGISREKVVPLYGRGSQ 85
> mmu:54197 Rnf5, 2410131O05Rik, AA407576, NG2; ring finger protein
5; K10666 E3 ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=180
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 47/82 (57%), Gaps = 5/82 (6%)
Query 41 SSSSSSSSRGATRKEGAS--PFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGD--- 95
+ G R+ G + FEC IC++ + VV+ CGHL+CW CLH WL+ D
Sbjct 4 AEEEDGGPEGPNRERGGASATFECNICLETAREAVVSVCGHLYCWPCLHQWLETRPDRQE 63
Query 96 CPICKAAVSADTVTPIYGRSSS 117
CP+CKA +S + V P+YGR S
Sbjct 64 CPVCKAGISREKVVPLYGRGSQ 85
> cel:C16C10.7 rnf-5; RiNg Finger protein family member (rnf-5);
K10666 E3 ubiquitin-protein ligase RNF5 [EC:6.3.2.19]
Length=235
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/104 (40%), Positives = 57/104 (54%), Gaps = 8/104 (7%)
Query 39 SNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGD--- 95
S + + S ++ K+ ++ FEC IC+D D VV+ CGHLFCW CL WL +
Sbjct 3 SETKAPSEEPTSSSNKDESARFECNICLDAAKDAVVSLCGHLFCWPCLSQWLDTRPNNQV 62
Query 96 CPICKAAVSADTVTPIYGR---SSSPKSHAHTRHFS--SETPPS 134
CP+CK+A+ + V PIYGR SS P+ R SE PP
Sbjct 63 CPVCKSAIDGNKVVPIYGRGGDSSDPRKKVPPRPKGQRSEPPPQ 106
> ath:AT4G03510 RMA1; RMA1; protein binding / ubiquitin-protein
ligase/ zinc ion binding
Length=249
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/68 (48%), Positives = 43/68 (63%), Gaps = 11/68 (16%)
Query 58 SPFECCICMDEIHDPVVTRCGHLFCWGCLHLWL-----------QQSGDCPICKAAVSAD 106
S F+C IC+D + +PVVT CGHLFCW C+H WL Q+ CP+CK+ VS
Sbjct 44 SNFDCNICLDSVQEPVVTLCGHLFCWPCIHKWLDVQSFSTSDEYQRHRQCPVCKSKVSHS 103
Query 107 TVTPIYGR 114
T+ P+YGR
Sbjct 104 TLVPLYGR 111
> xla:100049739 rnf5; ring finger protein 5; K10666 E3 ubiquitin-protein
ligase RNF5 [EC:6.3.2.19]
Length=168
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/82 (43%), Positives = 50/82 (60%), Gaps = 9/82 (10%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ---QSGDCPICKAAVSADTVTPIYGRSS 116
+EC IC++ +PVV+ CGHL+CW CLH WL+ + +CP+CKA VS + V PIYGR
Sbjct 12 YECNICLETAREPVVSVCGHLYCWPCLHQWLETRPERQECPVCKAGVSREKVIPIYGRGD 71
Query 117 SPKSHAHTRHFSSETPPSRPRA 138
+ + TPP RP+
Sbjct 72 GNQKDPRLK-----TPP-RPQG 87
> ath:AT4G28270 zinc finger (C3HC4-type RING finger) family protein
Length=193
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 52/94 (55%), Gaps = 25/94 (26%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGD----------------CPICKAAV 103
F+C IC+D++ DPVVT CGHLFCW C+H W S + CP+CK+ V
Sbjct 19 FDCNICLDQVRDPVVTLCGHLFCWPCIHKWTYASNNSRQRVDQYDHKREPPKCPVCKSDV 78
Query 104 SADTVTPIYGR-SSSPKSHAHTRHFSSETPPSRP 136
S T+ PIYGR +P+S ++ PSRP
Sbjct 79 SEATLVPIYGRGQKAPQSGSNV--------PSRP 104
> ath:AT2G44410 protein binding / zinc ion binding
Length=413
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 36/57 (63%), Gaps = 3/57 (5%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLH---LWLQQSGDCPICKAAVSADTVTPIYG 113
F+C IC+++ DP++T CGHLFCWGC + L +CP+C V+ V PIYG
Sbjct 123 FDCNICLEKAEDPILTCCGHLFCWGCFYQLPLIYLNIKECPVCDGEVTDAEVIPIYG 179
> ath:AT5G48655 zinc finger (C3HC4-type RING finger) family protein
Length=203
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 52/112 (46%), Gaps = 4/112 (3%)
Query 1 GGVASYCFCSSRPRSGCPSSSSSSNTIIIITSSSSSSNSNSSSSSSSSRGATRKEGASPF 60
GG + S R PSS S +I +S + N SS S S+ F
Sbjct 91 GGTTRFPANISNKRRRIPSSES----VIDCEHASVNDEVNMSSRVSRSKAPAPPPEEPKF 146
Query 61 ECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTPIY 112
C ICM + + T+CGH+FC GC+ + + + G CP C+ V+A + ++
Sbjct 147 TCPICMCPFTEEMSTKCGHIFCKGCIKMAISRQGKCPTCRKKVTAKELIRVF 198
> sce:YLR247C IRC20; Irc20p (EC:3.6.1.-)
Length=1556
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 0/60 (0%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTPIYGRSSSPK 119
C IC+ E+ + +CGH FC C+ WL+ CPICK S V ++S+ K
Sbjct 1237 LSCSICLGEVEIGAIIKCGHYFCKSCILTWLRAHSKCPICKGFCSISEVYNFKFKNSTEK 1296
> dre:100333470 ReO_6-like
Length=896
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 48/96 (50%), Gaps = 3/96 (3%)
Query 9 CSSRPRSGCPSSSSSSNTIIIITSSSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDE 68
C+SR + P+S SS II + + SS+ + + + FEC +CM
Sbjct 803 CTSRIK---PTSLLSSLNTIIPRDGTKAGYHVFSSAVVKRQVPLQLLDNADFECSLCMRL 859
Query 69 IHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVS 104
++PV T CGH FC CL L + +CP+CK +S
Sbjct 860 FYEPVTTPCGHTFCLKCLERCLDHNPNCPLCKENLS 895
> ath:AT5G01960 zinc finger (C3HC4-type RING finger) family protein
Length=426
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 48 SRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVS 104
S T + A+ C IC +++H P++ RC H+FC C+ W ++ CP+C+A V
Sbjct 351 SYATTEQVNAAGDLCAICQEKMHTPILLRCKHMFCEDCVSEWFERERTCPLCRALVK 407
> hsa:79836 LONRF3, FLJ22612, MGC119463, MGC119465, RNF127; LON
peptidase N-terminal domain and ring finger 3
Length=759
Score = 52.8 bits (125), Expect = 4e-07, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 57 ASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVS 104
AS EC +CM ++PV T CGH FC CL L + CP+CK +S
Sbjct 462 ASDLECALCMRLFYEPVTTPCGHTFCLKCLERCLDHNAKCPLCKDGLS 509
Score = 34.7 bits (78), Expect = 0.092, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSA 105
F+C C + DPV CGH FC CL C +C +SA
Sbjct 156 FKCRKCHGFLSDPVSLSCGHTFCKLCLERGRAADRRCALCGVKLSA 201
> ath:AT5G43420 zinc finger (C3HC4-type RING finger) family protein
Length=375
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 39/72 (54%), Gaps = 7/72 (9%)
Query 61 ECCICMDEIHDP----VVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTP---IYG 113
EC +C+ E D ++ C HLF C+ +WLQ + +CP+C+ VS DT P +
Sbjct 137 ECSVCLSEFQDEEKLRIIPNCSHLFHIDCIDVWLQNNANCPLCRTRVSCDTSFPPDRVSA 196
Query 114 RSSSPKSHAHTR 125
S+SP++ R
Sbjct 197 PSTSPENLVMLR 208
> cel:Y47G6A.31 hypothetical protein
Length=212
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 33/67 (49%), Gaps = 5/67 (7%)
Query 49 RGATRKEGASPFECCICMDEIHD-----PVVTRCGHLFCWGCLHLWLQQSGDCPICKAAV 103
RG E AS EC IC + D ++ CGH FC+ CL WL+ CP+C+ AV
Sbjct 21 RGVVSLEMASRLECSICFFDFDDFQHLPKLLENCGHTFCYSCLFDWLKSQDTCPMCREAV 80
Query 104 SADTVTP 110
+ P
Sbjct 81 NMTQEIP 87
> hsa:54941 RNF125, FLJ20456, MGC21737, TRAC1; ring finger protein
125; K12170 E3 ubiquitin-protein ligase RNF125 [EC:6.3.2.19]
Length=232
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 48/82 (58%), Gaps = 5/82 (6%)
Query 32 SSSSSSNSNSSSSSSSSRGATRKEG----ASPFECCICMDEIHDPVVTRCGHLFCWGCLH 87
S S+ + S+ +S+++R R+ + F+C +C++ +H PV TRCGH+FC C+
Sbjct 3 SVLSTDSGKSAPASATARALERRRDPELPVTSFDCAVCLEVLHQPVRTRCGHVFCRSCIA 62
Query 88 LWLQQSG-DCPICKAAVSADTV 108
L+ + CP C+A + ++ V
Sbjct 63 TSLKNNKWTCPYCRAYLPSEGV 84
> xla:443579 rnf219; ring finger protein 219
Length=659
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 59 PFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADT 107
P C IC+ ++ PV+ H+FC C+ LWL+ + CP+C+ ++A+
Sbjct 15 PITCHICLGKVRQPVICVNHHVFCTVCIELWLKNNNQCPVCRVPITAEN 63
> dre:414846 rnf141, zgc:86917; ring finger protein 141
Length=222
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 43/77 (55%), Gaps = 1/77 (1%)
Query 29 IITSSSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHL 88
+++S+ ++ S+ + +S R K+ ECCICMD D ++ C H FC C+
Sbjct 114 VLSSNVAAEGSSEDTCQASMRMGRVKQLTDEEECCICMDGKADLILP-CAHSFCQKCIDK 172
Query 89 WLQQSGDCPICKAAVSA 105
W QS +CP+C+ V+A
Sbjct 173 WSGQSRNCPVCRIQVTA 189
> hsa:164832 LONRF2, FLJ45273, MGC126711, MGC126713, RNF192; LON
peptidase N-terminal domain and ring finger 2
Length=754
Score = 52.0 bits (123), Expect = 6e-07, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 35/73 (47%), Gaps = 6/73 (8%)
Query 32 SSSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQ 91
S S NS + S S T FEC +CM + +PV T CGH FC CL L
Sbjct 425 SLQRSPNSETEESQGLSLDVT------DFECALCMRLLFEPVTTPCGHTFCLKCLERCLD 478
Query 92 QSGDCPICKAAVS 104
+ CP+CK +S
Sbjct 479 HAPHCPLCKDKLS 491
> mmu:381338 Lonrf2, 2900060P06Rik, AI851349; LON peptidase N-terminal
domain and ring finger 2
Length=518
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 57 ASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVT 109
A+ FEC +CM + +PV T CGH FC CL L + CP+CK +S T
Sbjct 208 ATDFECALCMRLLFEPVTTPCGHTFCLKCLERCLDHAPHCPLCKDKLSELLAT 260
> ath:AT2G26350 PEX10; PEX10; protein binding / zinc ion binding;
K13346 peroxin-10
Length=381
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 40/85 (47%), Gaps = 12/85 (14%)
Query 16 GCPSSSSSSNTIIIITSSSSSSNSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVT 75
G P + N I +S + N S+S S+S A K C +C+ P T
Sbjct 293 GLPVLNEEGNLI-----TSEAEKGNWSTSDSTSTEAVGK-------CTLCLSTRQHPTAT 340
Query 76 RCGHLFCWGCLHLWLQQSGDCPICK 100
CGH+FCW C+ W + +CP+C+
Sbjct 341 PCGHVFCWSCIMEWCNEKQECPLCR 365
> mmu:26374 Rfwd2, AI316802, C80879, Cop1; ring finger and WD
repeat domain 2; K10143 E3 ubiquitin-protein ligase RFWD2 [EC:6.3.2.19]
Length=733
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 54 KEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTPIY 112
++ ++ F C IC D I + +T+CGH FC+ C+H L+ + CP C V D + +Y
Sbjct 130 EDKSNDFVCPICFDMIEEAYMTKCGHSFCYKCIHQSLEDNNRCPKCNYVV--DNIDHLY 186
> mmu:74365 Lonrf3, 4932412G04Rik, 5730439E01Rik, A830039N02Rik,
AU023707, Rnf127; LON peptidase N-terminal domain and ring
finger 3
Length=753
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 21/44 (47%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 57 ASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICK 100
AS EC +CM ++PV T CGH FC CL L + CP+CK
Sbjct 456 ASDLECSLCMRLFYEPVTTPCGHTFCLKCLERCLDHNAKCPLCK 499
Score = 38.9 bits (89), Expect = 0.005, Method: Composition-based stats.
Identities = 26/91 (28%), Positives = 40/91 (43%), Gaps = 7/91 (7%)
Query 34 SSSSNSNSSSSSSSSRGATRKEGASP-----FECCICMDEIHDPVVTRCGHLFCWGCLHL 88
+S S+ SS +++ + A +P F+C C + DPV CGH FC CL
Sbjct 126 ASDSSGTSSCCAAALKEAGEAAAVAPEVWDGFKCKKCHGFLSDPVSLWCGHTFCKLCLER 185
Query 89 WLQQSGDCPICKAAVSADTVTPIYGRSSSPK 119
C +C +SA + GR+ P+
Sbjct 186 GRAADRRCALCGVKLSA--LMAASGRARGPR 214
> mmu:100504800 e3 ubiquitin-protein ligase RFWD2-like
Length=677
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 54 KEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTPIY 112
++ ++ F C IC D I + +T+CGH FC+ C+H L+ + CP C V D + +Y
Sbjct 74 EDKSNDFVCPICFDMIEEAYMTKCGHSFCYKCIHQSLEDNNRCPKCNYVV--DNIDHLY 130
> xla:779423 pdzrn3, MGC80196; PDZ domain containing ring finger
3
Length=1029
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSA 105
F+C +C + DP+ T CGH+FC GC+ W+ Q G CP+ +SA
Sbjct 16 FKCNLCNRVLEDPLTTPCGHVFCAGCVLPWVVQQGSCPVKCQRISA 61
> hsa:64326 RFWD2, COP1, FLJ10416, RNF200; ring finger and WD
repeat domain 2; K10143 E3 ubiquitin-protein ligase RFWD2 [EC:6.3.2.19]
Length=707
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 54 KEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTPIY 112
++ ++ F C IC D I + +T+CGH FC+ C+H L+ + CP C V D + +Y
Sbjct 128 EDKSNDFVCPICFDMIEEAYMTKCGHSFCYKCIHQSLEDNNRCPKCNYVV--DNIDHLY 184
> dre:100317496 si:dkey-258f14.5
Length=751
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAV 103
FEC +CM + PV T CGH FC CL L + CP+CK ++
Sbjct 455 FECSLCMRLFYQPVTTPCGHTFCTNCLERCLDHNPQCPLCKESL 498
Score = 29.3 bits (64), Expect = 4.0, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 24/63 (38%), Gaps = 4/63 (6%)
Query 38 NSNSSSSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCL-HLWLQQSGDC 96
N S S +R +C C I +PV CGH +C CL H Q C
Sbjct 84 NFKKKSEESPARSEPNWTQDCELDCPGCHCFIAEPVTVTCGHTYCRRCLQHSTFSQ---C 140
Query 97 PIC 99
+C
Sbjct 141 KVC 143
> pfa:PF10_0127 conserved Plasmodium protein
Length=520
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 0/62 (0%)
Query 53 RKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTVTPIY 112
++E F C +C+D PV+T C H+ C+ C++ L +CPICK + + + I
Sbjct 21 KEEIYENFICSVCLDLCDTPVITLCNHICCYKCMYYSLLHKRNCPICKQIIKHNNLKKIT 80
Query 113 GR 114
G+
Sbjct 81 GK 82
> xla:735224 hltf, MGC131155, smarca3; helicase-like transcription
factor; K15083 DNA repair protein RAD16
Length=999
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/87 (31%), Positives = 45/87 (51%), Gaps = 13/87 (14%)
Query 35 SSSNSNSSSSSSSSRGATRKE----------GASPFECCICMDEIHDPVVTRCGHLFCWG 84
SS+ S +S++ S+ G R++ S EC IC+D ++ PV+T C H+FC
Sbjct 713 SSTLSTMASTADSTPGDVREKLVQKIKLVLSSGSDEECAICLDSLNMPVITYCAHVFCKP 772
Query 85 CLHLWLQ---QSGDCPICKAAVSADTV 108
C+ +Q Q CP+C+ + D +
Sbjct 773 CICQVIQLKKQEAKCPLCRGLLRLDQL 799
> mmu:269695 Rnft2, AW049082, B830028P19Rik, Tmem118; ring finger
protein, transmembrane 2
Length=445
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 62 CCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSADTV 108
C IC E DP++ C H+FC CL LWL + CP+C+ +V+ DT+
Sbjct 385 CAICQAEFRDPMILLCQHVFCEECLCLWLDRERTCPLCR-SVAVDTL 430
> dre:393254 MGC56368, wu:fc04b02; zgc:56368
Length=479
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 39/76 (51%), Gaps = 10/76 (13%)
Query 43 SSSSSSRGATRKEGASPFECCICMDEIHDPVVTRCGHLFCWGCLHL-WLQQSGD-----C 96
+++ SRG +KE C IC+D DPV+ +CGH FC C+ + W + GD C
Sbjct 2 ATAGDSRGELQKE----LVCSICLDYFDDPVILKCGHNFCRMCILMHWEENGGDDVGYQC 57
Query 97 PICKAAVSADTVTPIY 112
P C+ + + T Y
Sbjct 58 PECRMVFAKMSFTKNY 73
> mmu:22670 Trim26, AI462198, Zfp173, Zfp1736; tripartite motif-containing
26; K12008 tripartite motif-containing protein
26
Length=545
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query 62 CCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGD---CPICKAAVSADTVTPIYGRSS 116
C IC+D + DPV CGH+FC C SG+ CP+CK + + P++ +S
Sbjct 16 CSICLDYLRDPVTIDCGHVFCRSCTSDIRPISGNRPVCPLCKKPFKKENIRPVWQLAS 73
> dre:564651 hltf; helicase-like transcription factor-like; K15083
DNA repair protein RAD16
Length=942
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 38/81 (46%), Gaps = 9/81 (11%)
Query 61 ECCICMDEIHDPVVTRCGHLFCWGCLHLWL---QQSGDCPICKAAVSADTVTPIYGRSSS 117
EC IC+D + PV+T C H+FC C+ + ++ CP+C+A + +
Sbjct 696 ECAICLDSLRQPVITYCAHVFCRPCICEVIRSEKEQAKCPLCRAQIKTKELV------EY 749
Query 118 PKSHAHTRHFSSETPPSRPRA 138
P A TR + E S +A
Sbjct 750 PGEQAETRSDTGENWRSSSKA 770
> hsa:91694 LONRF1, FLJ23749, RNF191; LON peptidase N-terminal
domain and ring finger 1
Length=773
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 58 SPFECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAV 103
S FEC +CM +PV T CGH FC CL L + CP+CK ++
Sbjct 475 SDFECSLCMRLFFEPVTTPCGHSFCKNCLERCLDHAPYCPLCKESL 520
> cel:ZC13.1 hypothetical protein
Length=398
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 62 CCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPICKAAVSAD 106
C +C ++ P+ C H+FC C+ WL Q CPIC+A V+ D
Sbjct 337 CAVCHGDLLQPIKLECTHVFCKFCIETWLDQKSTCPICRAEVTKD 381
> hsa:23024 PDZRN3, LNX3, SEMACAP3; PDZ domain containing ring
finger 3
Length=1066
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 31/53 (58%), Gaps = 1/53 (1%)
Query 60 FECCICMDEIHDPVVTRCGHLFCWGCLHLWLQQSGDCPI-CKAAVSADTVTPI 111
+C +C + DP+ T CGH+FC GC+ W+ Q G CP C+ +SA + +
Sbjct 16 LKCALCHKVLEDPLTTPCGHVFCAGCVLPWVVQEGSCPARCRGRLSAKELNHV 68
Lambda K H
0.315 0.125 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40