bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1239_orf1
Length=75
Score E
Sequences producing significant alignments: (Bits) Value
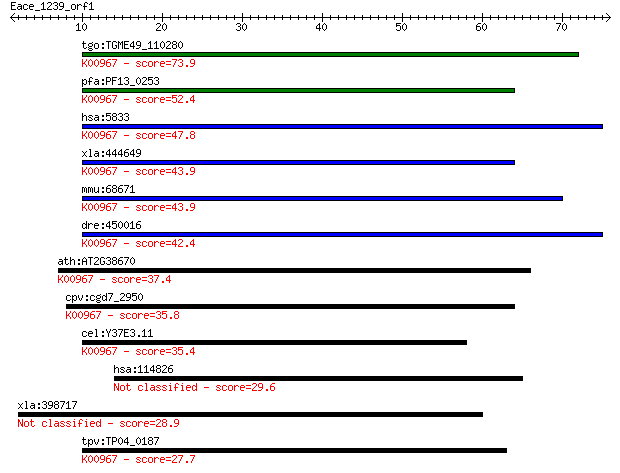
tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase, pu... 73.9 1e-13
pfa:PF13_0253 ethanolamine-phosphate cytidylyltransferase, put... 52.4 4e-07
hsa:5833 PCYT2, ET; phosphate cytidylyltransferase 2, ethanola... 47.8 9e-06
xla:444649 pcyt2, MGC84177; phosphate cytidylyltransferase 2, ... 43.9 1e-04
mmu:68671 Pcyt2, 1110033E03Rik, ET; phosphate cytidylyltransfe... 43.9 1e-04
dre:450016 pcyt2, im:7158585, wu:fb39h11, zgc:103434; phosphat... 42.4 4e-04
ath:AT2G38670 PECT1; PECT1 (PHOSPHORYLETHANOLAMINE CYTIDYLYLTR... 37.4 0.012
cpv:cgd7_2950 phospholipid cytidyltransferase HIGH family ; K0... 35.8 0.030
cel:Y37E3.11 hypothetical protein; K00967 ethanolamine-phospha... 35.4 0.047
hsa:114826 SMYD4, KIAA1936, ZMYND21; SET and MYND domain conta... 29.6 2.2
xla:398717 invs-b, NPH2, NPHP2; inversin 28.9 4.6
tpv:TP04_0187 ethanolamine-phosphate cytidylyltransferase (EC:... 27.7 9.1
> tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase,
putative (EC:4.1.1.70 2.7.7.39 2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=1128
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/63 (57%), Positives = 42/63 (66%), Gaps = 1/63 (1%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFW-ESLSGQT 68
DPY+VPKE+G+YRE ES S WTTR LV R++ NR+ L ATI R KE FW E GQ
Sbjct 1063 DPYRVPKELGVYREVESSSSWTTRALVERILANREALMATIETRCSKEAKFWREQEQGQM 1122
Query 69 ARL 71
L
Sbjct 1123 VSL 1125
> pfa:PF13_0253 ethanolamine-phosphate cytidylyltransferase, putative
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=573
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 22/54 (40%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWES 63
DPY +PK++ IY+E S S TT E++ R+ +N++ L + R KKE WE+
Sbjct 512 DPYDIPKKLNIYQELSSESNITTYEIIQRIEKNKKYLMRNMSKRNKKEESIWET 565
> hsa:5833 PCYT2, ET; phosphate cytidylyltransferase 2, ethanolamine
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=407
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 26/65 (40%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWESLSGQTA 69
DPY+ PK GI+R+ +S S TT +V R+I NR A +E KE F E+ Q A
Sbjct 338 DPYQEPKRRGIFRQIDSGSNLTTDLIVQRIITNRLEYEARNQKKEAKELAFLEAARQQAA 397
Query 70 RLIAE 74
+ + E
Sbjct 398 QPLGE 402
> xla:444649 pcyt2, MGC84177; phosphate cytidylyltransferase 2,
ethanolamine (EC:2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=383
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWES 63
DPY PK+ GI+R +S + TT ++V R+I+NR A +E KE +E+
Sbjct 316 DPYAEPKQRGIFRAVDSGNSLTTDDIVQRIIKNRLEYEARNQKKEAKELAVFEA 369
> mmu:68671 Pcyt2, 1110033E03Rik, ET; phosphate cytidylyltransferase
2, ethanolamine (EC:2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=404
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWESLSGQTA 69
DPY+ PK GI+ + +S S TT +V R+I+NR A +E KE F E+ Q A
Sbjct 338 DPYQEPKRRGIFYQIDSGSDLTTDLIVQRIIKNRLEYEARNQKKEAKELAFLEATKQQEA 397
> dre:450016 pcyt2, im:7158585, wu:fb39h11, zgc:103434; phosphate
cytidylyltransferase 2, ethanolamine (EC:2.7.7.14); K00967
ethanolamine-phosphate cytidylyltransferase [EC:2.7.7.14]
Length=397
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWESLSGQTA 69
DPY PK+ GI+R +S + TT ++V R+I+NR A +E KE ++L +
Sbjct 327 DPYAEPKKRGIFRILDSSNNLTTDDIVQRIIENRLQFEARNQKKEAKEMAVLQALKRKDE 386
Query 70 RLIAE 74
+ +E
Sbjct 387 SVKSE 391
> ath:AT2G38670 PECT1; PECT1 (PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE
1); ethanolamine-phosphate cytidylyltransferase
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=421
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 7 QQEDPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWESLS 65
++++PY VP MGI++ +S TT ++ R++ N + +E E ++E S
Sbjct 358 EEDNPYSVPISMGIFQVLDSPLDITTSTIIRRIVANHEAYQKRNAKKEASEKKYYEQKS 416
> cpv:cgd7_2950 phospholipid cytidyltransferase HIGH family ;
K00967 ethanolamine-phosphate cytidylyltransferase [EC:2.7.7.14]
Length=405
Score = 35.8 bits (81), Expect = 0.030, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query 8 QEDPYKVPKEMGIYREA---ESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWES 63
+E Y++PK+MGI+ + + T R++ SR++ R ++ I R KE F+E+
Sbjct 340 EEFCYEIPKKMGIFCNTTILKKEEICTNRDIFSRIMNRRDSILYVIKKRWAKELSFYEN 398
> cel:Y37E3.11 hypothetical protein; K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=377
Score = 35.4 bits (80), Expect = 0.047, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKE 57
DP+ K GIY E +S S TT ++ R+I +R +EKKE
Sbjct 320 DPFAEAKRRGIYHEVDSGSDMTTDLIIDRIIHHRLEYETRNKKKEKKE 367
> hsa:114826 SMYD4, KIAA1936, ZMYND21; SET and MYND domain containing
4
Length=804
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 1/51 (1%)
Query 14 VPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWESL 64
V + G R+AESF LW +V + A +G+ +K TH SL
Sbjct 677 VQRLSGCQRDAESF-LWAEHAVVGEIADGLARACAALGDWQKSATHLQRSL 726
> xla:398717 invs-b, NPH2, NPHP2; inversin
Length=1002
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 2 GGGFIQQEDPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETH 59
G G + Q + KV +GI + + E SRV + R+T+SA G R ETH
Sbjct 819 GCGKLSQSE--KVSLGIGIQGRVDCITSPEPCETPSRVCRERKTISAKTGQRPLTETH 874
> tpv:TP04_0187 ethanolamine-phosphate cytidylyltransferase (EC:2.7.7.14);
K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=385
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 10 DPYKVPKEMGIYREAESFSLWTTRELVSRVIQNRQTLSATIGNREKKETHFWE 62
+P +V + +GI R +S T+ E++ RV + + +R KKE + ++
Sbjct 330 NPLEVVESLGILRYVDSGLKTTSSEIIKRVSDRMGQIRRNVSDRCKKELNHYK 382
Lambda K H
0.313 0.130 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2002740660
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40