bitscore colors: <40, 40-50 , 50-80, 80-200, >200
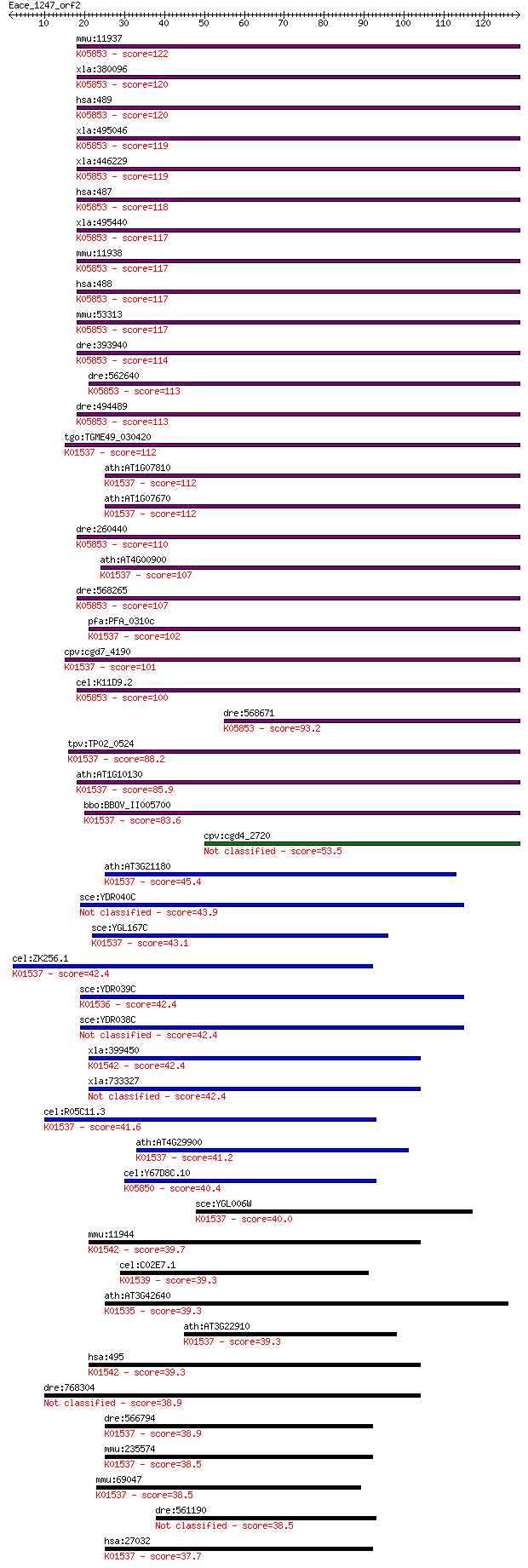
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1247_orf2
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
mmu:11937 Atp2a1, SERCA1; ATPase, Ca++ transporting, cardiac m... 122 3e-28
xla:380096 atp2a2, MGC52615, atp2b, ca-p60a, dar, serca2; ATPa... 120 1e-27
hsa:489 ATP2A3, SERCA3; ATPase, Ca++ transporting, ubiquitous ... 120 1e-27
xla:495046 hypothetical LOC495046; K05853 Ca2+ transporting AT... 119 3e-27
xla:446229 atp2a1, atp2a, atp2a2, serca1; ATPase, Ca++ transpo... 119 3e-27
hsa:487 ATP2A1, ATP2A, SERCA1; ATPase, Ca++ transporting, card... 118 5e-27
xla:495440 atp2a3, serca3; ATPase, Ca++ transporting, ubiquito... 117 8e-27
mmu:11938 Atp2a2, 9530097L16Rik, D5Wsu150e, KIAA4195, SERCA2, ... 117 9e-27
hsa:488 ATP2A2, ATP2B, DAR, DD, DKFZp686P0211, FLJ20293, FLJ38... 117 9e-27
mmu:53313 Atp2a3, SERCA3b, Serca3; ATPase, Ca++ transporting, ... 117 1e-26
dre:393940 atp2a2a, MGC55380, atp2a2, zgc:55380; ATPase, Ca++ ... 114 6e-26
dre:562640 si:dkey-28b4.8; K05853 Ca2+ transporting ATPase, sa... 113 1e-25
dre:494489 atp2a1l, zgc:154094; ATPase, Ca++ transporting, car... 113 2e-25
tgo:TGME49_030420 calcium-transporting ATPase, putative (EC:3.... 112 3e-25
ath:AT1G07810 ECA1; ECA1 (ER-TYPE CA2+-ATPASE 1); calcium-tran... 112 3e-25
ath:AT1G07670 calcium-transporting ATPase; K01537 Ca2+-transpo... 112 3e-25
dre:260440 atp2a1, MGC92110, cb279, serca, serca1, wu:cegs655,... 110 7e-25
ath:AT4G00900 ECA2; ECA2 (ER-TYPE CA2+-ATPASE 2); calcium-tran... 107 8e-24
dre:568265 atp2a2b, SERCA2, fb51e04, fb80c02, si:dkey-83d9.6, ... 107 1e-23
pfa:PFA_0310c calcium-transporting ATPase, putative (EC:3.6.3.... 102 2e-22
cpv:cgd7_4190 cation-transporting P-type ATpase with 11 or mor... 101 5e-22
cel:K11D9.2 sca-1; SERCA (Sarco-Endoplasmic Reticulum Calcium ... 100 9e-22
dre:568671 atp2a3, si:dkey-205l20.1; ATPase, Ca++ transporting... 93.2 2e-19
tpv:TP02_0524 calcium-transporting ATPase; K01537 Ca2+-transpo... 88.2 6e-18
ath:AT1G10130 ECA3; ECA3 (ENDOPLASMIC RETICULUM-TYPE CALCIUM-T... 85.9 3e-17
bbo:BBOV_II005700 18.m06474; calcium ATPase SERCA-like (EC:3.6... 83.6 1e-16
cpv:cgd4_2720 P-type ATpase involved in cation transport 53.5 1e-07
ath:AT3G21180 ACA9; ACA9 (AUTOINHIBITED CA(2+)-ATPASE 9); calc... 45.4 4e-05
sce:YDR040C ENA1, HOR6, PMR2; Ena1p (EC:3.6.3.7) 43.9 1e-04
sce:YGL167C PMR1, BSD1, LDB1, SSC1; High affinity Ca2+/Mn2+ P-... 43.1 2e-04
cel:ZK256.1 pmr-1; PMR-type Golgi ATPase family member (pmr-1)... 42.4 3e-04
sce:YDR039C ENA2; Ena2p (EC:3.6.3.7 3.6.3.12); K01536 Na+-expo... 42.4 4e-04
sce:YDR038C ENA5; Ena5p (EC:3.6.3.7) 42.4 4e-04
xla:399450 atp4a; ATPase, H+/K+ exchanging, alpha polypeptide ... 42.4 4e-04
xla:733327 hypothetical protein LOC733327 42.4 4e-04
cel:R05C11.3 ATPase; hypothetical protein; K01537 Ca2+-transpo... 41.6 6e-04
ath:AT4G29900 ACA10; ACA10 (AUTOINHIBITED CA(2+)-ATPASE 10); c... 41.2 9e-04
cel:Y67D8C.10 mca-3; Membrane Calcium ATPase family member (mc... 40.4 0.001
sce:YGL006W PMC1; Pmc1p (EC:3.6.3.8); K01537 Ca2+-transporting... 40.0 0.002
mmu:11944 Atp4a; ATPase, H+/K+ exchanging, gastric, alpha poly... 39.7 0.002
cel:C02E7.1 hypothetical protein; K01539 sodium/potassium-tran... 39.3 0.003
ath:AT3G42640 AHA8; AHA8 (Arabidopsis H(+)-ATPase 8); ATPase; ... 39.3 0.003
ath:AT3G22910 calcium-transporting ATPase, plasma membrane-typ... 39.3 0.003
hsa:495 ATP4A, ATP6A; ATPase, H+/K+ exchanging, alpha polypept... 39.3 0.003
dre:768304 atp2b4, MGC152801, zgc:152801; ATPase, Ca++ transpo... 38.9 0.004
dre:566794 atp2c1; ATPase, Ca++ transporting, type 2C, member ... 38.9 0.004
mmu:235574 Atp2c1, 1700121J11Rik, ATP2C1A, AW061228, BCPM, D93... 38.5 0.005
mmu:69047 Atp2c2, 1810010G06Rik, KIAA0703, Spca2, mKIAA0703; A... 38.5 0.006
dre:561190 atp2b1b; ATPase, Ca++ transporting, plasma membrane 1b 38.5 0.006
hsa:27032 ATP2C1, ATP2C1A, BCPM, HHD, KIAA1347, PMR1, SPCA1, h... 37.7 0.010
> mmu:11937 Atp2a1, SERCA1; ATPase, Ca++ transporting, cardiac
muscle, fast twitch 1 (EC:3.6.3.8); K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=994
Score = 122 bits (306), Expect = 3e-28, Method: Composition-based stats.
Identities = 63/111 (56%), Positives = 79/111 (71%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
EA H+ + +E L Y S +GL + V LE++GPNEL EEGKSL++L++EQF+DLL
Sbjct 2 EAAHSKSTEECLSYFGVSETTGLTPDQVKRHLEKYGPNELPAEEGKSLWELVVEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETVTAFVEPFVILLILIANAIVGVWQERNA 112
> xla:380096 atp2a2, MGC52615, atp2b, ca-p60a, dar, serca2; ATPase,
Ca++ transporting, cardiac muscle, slow twitch 2; K05853
Ca2+ transporting ATPase, sarcoplasmic/endoplasmic reticulum
[EC:3.6.3.8]
Length=996
Score = 120 bits (301), Expect = 1e-27, Method: Composition-based stats.
Identities = 61/111 (54%), Positives = 77/111 (69%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E H T +E L Y + +GL E+V + E++GPNEL EEGKS+++L+ EQF+DLL
Sbjct 2 ENAHAKTTEECLAYFGVNENTGLSPEIVKKNFEKYGPNELPAEEGKSIWELVAEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETVTAFVEPFVILLILIANAVVGVWQERNA 112
> hsa:489 ATP2A3, SERCA3; ATPase, Ca++ transporting, ubiquitous
(EC:3.6.3.8); K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic
reticulum [EC:3.6.3.8]
Length=999
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 67/112 (59%), Positives = 80/112 (71%), Gaps = 2/112 (1%)
Query 18 EAPHTCTVQEVLEYHETSLESGL-PAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDL 76
EA H +VL + + E GL PA+V R ER+GPNEL EEGKSL++L+LEQF+DL
Sbjct 2 EAAHLLPAADVLRHFSVTAEGGLSPAQVTGAR-ERYGPNELPSEEGKSLWELVLEQFEDL 60
Query 77 LVRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
LVRILL AA+VSFVLA E GE A+VEPLVI++IL NA V VWQE NA
Sbjct 61 LVRILLLAALVSFVLAWFEEGEETTTAFVEPLVIMLILVANAIVGVWQERNA 112
> xla:495046 hypothetical LOC495046; K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1042
Score = 119 bits (297), Expect = 3e-27, Method: Composition-based stats.
Identities = 63/111 (56%), Positives = 80/111 (72%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT TV+EVL + + +GL E V ++ ER+G NEL EEGK+L++L++EQF+DLL
Sbjct 2 ENAHTKTVEEVLAHFNVNESTGLSLEQVKKQKERWGTNELPAEEGKTLWELVIEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETVTAFVEPFVILLILVANAIVGVWQERNA 112
> xla:446229 atp2a1, atp2a, atp2a2, serca1; ATPase, Ca++ transporting,
cardiac muscle, fast twitch 1 (EC:3.6.3.8); K05853
Ca2+ transporting ATPase, sarcoplasmic/endoplasmic reticulum
[EC:3.6.3.8]
Length=1042
Score = 119 bits (297), Expect = 3e-27, Method: Composition-based stats.
Identities = 63/111 (56%), Positives = 80/111 (72%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT TV+EVL + + +GL E V ++ ER+G NEL EEGK+L++L++EQF+DLL
Sbjct 2 ENAHTKTVEEVLAHCNVNESTGLSLEQVKKQKERWGTNELPAEEGKTLWELVVEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNA 112
> hsa:487 ATP2A1, ATP2A, SERCA1; ATPase, Ca++ transporting, cardiac
muscle, fast twitch 1 (EC:3.6.3.8); K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=994
Score = 118 bits (295), Expect = 5e-27, Method: Composition-based stats.
Identities = 62/111 (55%), Positives = 77/111 (69%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
EA H T +E L Y S +GL + V LE++G NEL EEGK+L++L++EQF+DLL
Sbjct 2 EAAHAKTTEECLAYFGVSETTGLTPDQVKRNLEKYGLNELPAEEGKTLWELVIEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILIANAIVGVWQERNA 112
> xla:495440 atp2a3, serca3; ATPase, Ca++ transporting, ubiquitous
(EC:3.6.3.8); K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic
reticulum [EC:3.6.3.8]
Length=1033
Score = 117 bits (293), Expect = 8e-27, Method: Composition-based stats.
Identities = 63/111 (56%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
++ H TV EVL E + GL +E V E++G NEL EEGKSL++L+LEQF+DLL
Sbjct 2 DSAHAKTVTEVLRLFEVNENCGLSSEQVRRSREKYGLNELPAEEGKSLWELVLEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA VSFVLA E GE A+VEP+VI++IL +NA V VWQE NA
Sbjct 62 VRILLLAAFVSFVLAWFEEGEETTTAFVEPIVIIMILVINAFVGVWQERNA 112
> mmu:11938 Atp2a2, 9530097L16Rik, D5Wsu150e, KIAA4195, SERCA2,
SERCA2B, mKIAA4195; ATPase, Ca++ transporting, cardiac muscle,
slow twitch 2 (EC:3.6.3.8); K05853 Ca2+ transporting ATPase,
sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1044
Score = 117 bits (293), Expect = 9e-27, Method: Composition-based stats.
Identities = 63/111 (56%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT TV+EVL + + +GL E V + ER+G NEL EEGK+L +L++EQF+DLL
Sbjct 2 ENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNA 112
> hsa:488 ATP2A2, ATP2B, DAR, DD, DKFZp686P0211, FLJ20293, FLJ38063,
MGC45367, SERCA2; ATPase, Ca++ transporting, cardiac
muscle, slow twitch 2 (EC:3.6.3.8); K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1015
Score = 117 bits (293), Expect = 9e-27, Method: Composition-based stats.
Identities = 63/111 (56%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT TV+EVL + + +GL E V + ER+G NEL EEGK+L +L++EQF+DLL
Sbjct 2 ENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNA 112
> mmu:53313 Atp2a3, SERCA3b, Serca3; ATPase, Ca++ transporting,
ubiquitous (EC:3.6.3.8); K05853 Ca2+ transporting ATPase,
sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=999
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 63/111 (56%), Positives = 77/111 (69%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E H + +VL + E GL E V + ER+GPNEL EEGKSL++L++EQF+DLL
Sbjct 2 EEAHLLSAADVLRRFSVTAEGGLSLEQVTDARERYGPNELPTEEGKSLWELVVEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA+VSFVLA E GE A+VEPLVI++IL NA V VWQE NA
Sbjct 62 VRILLLAALVSFVLAWFEEGEETTTAFVEPLVIMLILVANAIVGVWQERNA 112
> dre:393940 atp2a2a, MGC55380, atp2a2, zgc:55380; ATPase, Ca++
transporting, cardiac muscle, slow twitch 2a (EC:3.6.3.8);
K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic reticulum
[EC:3.6.3.8]
Length=996
Score = 114 bits (286), Expect = 6e-26, Method: Composition-based stats.
Identities = 59/111 (53%), Positives = 77/111 (69%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT +V+EV + +GL + V +++GPNEL EEGKS+++L++EQF+DLL
Sbjct 2 ENAHTKSVEEVYSNFSVNESTGLTLDQVKRNRDKWGPNELPAEEGKSIWELVIEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILIANAIVGVWQERNA 112
> dre:562640 si:dkey-28b4.8; K05853 Ca2+ transporting ATPase,
sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1050
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 61/108 (56%), Positives = 76/108 (70%), Gaps = 6/108 (5%)
Query 21 HTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRI 80
HT TV+EVL Y + +GL +E + + ER+GPN GKSL++L+LEQF+DLLVRI
Sbjct 5 HTKTVEEVLGYFSVNETTGLSSEQLRKSRERWGPN------GKSLWELVLEQFEDLLVRI 58
Query 81 LLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
LL AA +SF LA E GEG + A+VEP VIL+IL NA V VWQE NA
Sbjct 59 LLLAACISFTLAWFEEGEGTITAFVEPFVILLILIANAIVGVWQERNA 106
> dre:494489 atp2a1l, zgc:154094; ATPase, Ca++ transporting, cardiac
muscle, fast twitch 1 like; K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=991
Score = 113 bits (282), Expect = 2e-25, Method: Composition-based stats.
Identities = 61/111 (54%), Positives = 74/111 (66%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT E L Y S +GL E + L ++G NEL EEGKS+++LI+EQF+DLL
Sbjct 2 EDAHTKGPAECLAYFSVSETTGLSPEQFKKNLAKYGYNELPAEEGKSIWELIIEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILIANAVVGVWQERNA 112
> tgo:TGME49_030420 calcium-transporting ATPase, putative (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1093
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 69/115 (60%), Positives = 83/115 (72%), Gaps = 1/115 (0%)
Query 15 SLQEAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQ 74
++ H +EV+ + + GL ERLE FG NEL+ E GKSL QLILEQFQ
Sbjct 40 NIHTKAHVLDAEEVVRQLKADAKRGLSEADACERLELFGKNELEQEPGKSLLQLILEQFQ 99
Query 75 DLLVRILLAAAVVSFVLAVVEGG-EGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
DLLVRILL+AAVVSF+LA+ EGG + G+ A++EPLVILIIL LNAAV VWQESNA
Sbjct 100 DLLVRILLSAAVVSFILALFEGGADEGVTAFIEPLVILIILILNAAVGVWQESNA 154
> ath:AT1G07810 ECA1; ECA1 (ER-TYPE CA2+-ATPASE 1); calcium-transporting
ATPase; K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1061
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 60/107 (56%), Positives = 77/107 (71%), Gaps = 3/107 (2%)
Query 25 VQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAA 84
V E E+ S E GL ++ V +R + +G NEL+ EG S+F+LILEQF D LVRILLAA
Sbjct 31 VAECEEHFVVSREKGLSSDEVLKRHQIYGLNELEKPEGTSIFKLILEQFNDTLVRILLAA 90
Query 85 AVVSFVLAVVEGGEG---GLAAYVEPLVILIILFLNAAVXVWQESNA 128
AV+SFVLA +G EG G+ A+VEPLVI +IL +NA V +WQE+NA
Sbjct 91 AVISFVLAFFDGDEGGEMGITAFVEPLVIFLILIVNAIVGIWQETNA 137
> ath:AT1G07670 calcium-transporting ATPase; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1061
Score = 112 bits (280), Expect = 3e-25, Method: Composition-based stats.
Identities = 61/107 (57%), Positives = 75/107 (70%), Gaps = 3/107 (2%)
Query 25 VQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAA 84
V E E S E GL + V +R + +G NEL+ EG S+F+LILEQF D LVRILLAA
Sbjct 31 VSECEEKFGVSREKGLSTDEVLKRHQIYGLNELEKPEGTSIFKLILEQFNDTLVRILLAA 90
Query 85 AVVSFVLAVV---EGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
AV+SFVLA EGGE G+ A+VEPLVI +IL +NA V +WQE+NA
Sbjct 91 AVISFVLAFFDGDEGGEMGITAFVEPLVIFLILIVNAIVGIWQETNA 137
> dre:260440 atp2a1, MGC92110, cb279, serca, serca1, wu:cegs655,
wu:fb17h11, wu:fb19b10, zgc:92110; ATPase, Ca++ transporting,
cardiac muscle, fast twitch 1 (EC:3.6.3.8); K05853 Ca2+
transporting ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=994
Score = 110 bits (276), Expect = 7e-25, Method: Composition-based stats.
Identities = 60/111 (54%), Positives = 74/111 (66%), Gaps = 0/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT E L Y S +GL E V + ++G NEL EEGKS+++L++EQF+DLL
Sbjct 2 ENAHTKETPECLAYFGVSESTGLTPEQVKKNQAKYGFNELPAEEGKSIWELVVEQFEDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 62 VRILLLAACISFVLAWFEEGEETVTAFVEPFVILLILIANAVVGVWQERNA 112
> ath:AT4G00900 ECA2; ECA2 (ER-TYPE CA2+-ATPASE 2); calcium-transporting
ATPase; K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1054
Score = 107 bits (268), Expect = 8e-24, Method: Composition-based stats.
Identities = 58/108 (53%), Positives = 78/108 (72%), Gaps = 3/108 (2%)
Query 24 TVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLA 83
+V++ L+ ++T L+ GL +E V R +++G NEL E+GK L+ L+LEQF D LV+ILL
Sbjct 13 SVEQCLKEYKTRLDKGLTSEDVQIRRQKYGFNELAKEKGKPLWHLVLEQFDDTLVKILLG 72
Query 84 AAVVSFVLAVV--EGGEG-GLAAYVEPLVILIILFLNAAVXVWQESNA 128
AA +SFVLA + E G G G A+VEP VI++IL LNA V VWQESNA
Sbjct 73 AAFISFVLAFLGEEHGSGSGFEAFVEPFVIVLILILNAVVGVWQESNA 120
> dre:568265 atp2a2b, SERCA2, fb51e04, fb80c02, si:dkey-83d9.6,
wu:fb51e04, wu:fb80c02, zK83D9.6; ATPase, Ca++ transporting,
cardiac muscle, slow twitch 2b; K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1035
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 59/111 (53%), Positives = 75/111 (67%), Gaps = 6/111 (5%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E HT TV+EV + + +GL E V + E++GPN GKSL++L++EQF+DLL
Sbjct 2 ENAHTKTVEEVYSFFAVNESTGLGLEQVKRQREKWGPN------GKSLWELVVEQFEDLL 55
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
VRILL AA +SFVLA E GE + A+VEP VIL+IL NA V VWQE NA
Sbjct 56 VRILLLAACISFVLAWFEEGEETITAFVEPFVILLILIANAIVGVWQERNA 106
> pfa:PFA_0310c calcium-transporting ATPase, putative (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1228
Score = 102 bits (255), Expect = 2e-22, Method: Composition-based stats.
Identities = 53/110 (48%), Positives = 79/110 (71%), Gaps = 2/110 (1%)
Query 21 HTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRI 80
HT V++VL++ + + ++GL E + +R ++G NEL+ E+ KS+F+LIL QF DLLV+I
Sbjct 9 HTYDVEDVLKFLDVNKDNGLKNEELDDRRLKYGLNELEVEKKKSIFELILNQFDDLLVKI 68
Query 81 LLAAAVVSFVLAVVEGGEGG--LAAYVEPLVILIILFLNAAVXVWQESNA 128
LL AA +SFVL +++ + ++EPLVI++IL LNAAV VWQE NA
Sbjct 69 LLLAAFISFVLTLLDMKHKKIEICDFIEPLVIVLILILNAAVGVWQECNA 118
> cpv:cgd7_4190 cation-transporting P-type ATpase with 11 or more
transmembrane domains ; K01537 Ca2+-transporting ATPase
[EC:3.6.3.8]
Length=1129
Score = 101 bits (252), Expect = 5e-22, Method: Composition-based stats.
Identities = 55/116 (47%), Positives = 73/116 (62%), Gaps = 2/116 (1%)
Query 15 SLQEAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQ 74
SL E PH + E+L ++ + GL V + + FG N L+ E S + LIL QF
Sbjct 3 SLLEDPHVKSCDEILRHYNVDCDVGLSNGQVEQYTQLFGKNSLEEPEKTSYWALILAQFD 62
Query 75 DLLVRILLAAAVVSFVLAVV--EGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
DLLVRILL AA++SF A++ E G++A++EP+VIL IL LNA V VWQESNA
Sbjct 63 DLLVRILLGAALMSFFFALIGDNAYEEGISAFIEPIVILFILVLNAFVGVWQESNA 118
> cel:K11D9.2 sca-1; SERCA (Sarco-Endoplasmic Reticulum Calcium
ATPase) family member (sca-1); K05853 Ca2+ transporting ATPase,
sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1004
Score = 100 bits (250), Expect = 9e-22, Method: Composition-based stats.
Identities = 58/114 (50%), Positives = 73/114 (64%), Gaps = 4/114 (3%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E H EV ++ T E P +V R ++G NE+ EEGKSL++LILEQF DLL
Sbjct 2 EDAHAKDANEVCKFFGTGPEGLTPQQVETLR-NKYGENEMPAEEGKSLWELILEQFDDLL 60
Query 78 VRILLAAAVVSFVLAVVEGGEG---GLAAYVEPLVILIILFLNAAVXVWQESNA 128
V+ILL AA++SFVLA+ E E + A+VEP VIL+IL NA V VWQE NA
Sbjct 61 VKILLLAAIISFVLALFEEHEDQTEAVTAFVEPFVILLILIANATVGVWQERNA 114
> dre:568671 atp2a3, si:dkey-205l20.1; ATPase, Ca++ transporting,
ubiquitous; K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic
reticulum [EC:3.6.3.8]
Length=970
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/74 (64%), Positives = 58/74 (78%), Gaps = 0/74 (0%)
Query 55 NELQHEEGKSLFQLILEQFQDLLVRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIIL 114
EL EEGKSL++L++EQF+DLLVRILL AA VSFVLA+ E GE A+VEP+VIL+IL
Sbjct 4 TELPAEEGKSLWELVVEQFEDLLVRILLLAACVSFVLALFEEGEESTTAFVEPIVILLIL 63
Query 115 FLNAAVXVWQESNA 128
NA + VWQE NA
Sbjct 64 VANAVIGVWQERNA 77
> tpv:TP02_0524 calcium-transporting ATPase; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1277
Score = 88.2 bits (217), Expect = 6e-18, Method: Composition-based stats.
Identities = 48/114 (42%), Positives = 68/114 (59%), Gaps = 1/114 (0%)
Query 16 LQEAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQD 75
L +PH EVL ++ +L+ GL E V E G + + SL L ++QF D
Sbjct 7 LLASPHVYESHEVLNHYSVNLDYGLNDEQVLLHREVLGSHSFLKPKKLSLLHLFIQQFDD 66
Query 76 LLVRILLAAAVVSFVLAVVEGGEGG-LAAYVEPLVILIILFLNAAVXVWQESNA 128
LLV+ILL+AA+VSF + E +++++EP+VIL IL LNA V VWQE+NA
Sbjct 67 LLVKILLSAAIVSFFFTCFDPHEKKHISSFIEPIVILFILILNALVGVWQEANA 120
> ath:AT1G10130 ECA3; ECA3 (ENDOPLASMIC RETICULUM-TYPE CALCIUM-TRANSPORTING
ATPASE 3); calcium-transporting ATPase/ calmodulin
binding / manganese-transporting ATPase/ peroxidase; K01537
Ca2+-transporting ATPase [EC:3.6.3.8]
Length=998
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 50/111 (45%), Positives = 68/111 (61%), Gaps = 1/111 (0%)
Query 18 EAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLL 77
E + +V EVL++ GL V +G N L E+ ++L+L+QF DLL
Sbjct 2 EDAYARSVSEVLDFFGVDPTKGLSDSQVVHHSRLYGRNVLPEEKRTPFWKLVLKQFDDLL 61
Query 78 VRILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQESNA 128
V+IL+ AA+VSFVLA+ GE GL A++EP VIL+IL NAAV V E+NA
Sbjct 62 VKILIVAAIVSFVLALAN-GETGLTAFLEPFVILLILAANAAVGVITETNA 111
> bbo:BBOV_II005700 18.m06474; calcium ATPase SERCA-like (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1028
Score = 83.6 bits (205), Expect = 1e-16, Method: Composition-based stats.
Identities = 55/110 (50%), Positives = 77/110 (70%), Gaps = 1/110 (0%)
Query 20 PHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVR 79
PHT +V +VL+++ +L+ GL ++ V RL+++GPN L +SL L + QF DLLV+
Sbjct 19 PHTTSVDDVLKHYGVTLQHGLDSKTVELRLKQYGPNMLAQHSKESLLSLFISQFDDLLVK 78
Query 80 ILLAAAVVSFVLAVVEGGEG-GLAAYVEPLVILIILFLNAAVXVWQESNA 128
ILL AAV+SF+L + E E + ++EPLVIL+IL LNA V VWQESNA
Sbjct 79 ILLGAAVISFILTLTEVSESYAITDFIEPLVILLILILNAIVGVWQESNA 128
> cpv:cgd4_2720 P-type ATpase involved in cation transport
Length=1528
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 50/79 (63%), Gaps = 10/79 (12%)
Query 50 ERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAAAVVSFVLAVVEGGEGGLAAYVEPLV 109
+++GPN+LQ E+ + ++++ L QF L+V +LL A +VS LA E +VE +
Sbjct 245 KKYGPNKLQKEKHEPIWRIFLSQFTSLVVSLLLVAGIVS--LAFQE--------WVEGIG 294
Query 110 ILIILFLNAAVXVWQESNA 128
IL I+ +NA++ + E+NA
Sbjct 295 ILFIVLINASLATYMENNA 313
> ath:AT3G21180 ACA9; ACA9 (AUTOINHIBITED CA(2+)-ATPASE 9); calcium-transporting
ATPase/ calmodulin binding; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1086
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 51/96 (53%), Gaps = 8/96 (8%)
Query 25 VQEVLEYHETSLESGLPAEV--VAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILL 82
V+ V E ++++E G+ + V +R FG N ++GK+ F + E +QDL + IL+
Sbjct 153 VKGVAEKLKSNMEQGINEDEKEVIDRKNAFGSNTYPKKKGKNFFMFLWEAWQDLTLIILI 212
Query 83 AAAVVSFVLAVVEGG------EGGLAAYVEPLVILI 112
AAV S L + G +GG A+ LVI++
Sbjct 213 IAAVTSLALGIKTEGLKEGWLDGGSIAFAVLLVIVV 248
> sce:YDR040C ENA1, HOR6, PMR2; Ena1p (EC:3.6.3.7)
Length=1091
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 50/96 (52%), Gaps = 2/96 (2%)
Query 19 APHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLV 78
A HT T +E E+ TSL GL + RL+ G N L + ++L Q + ++
Sbjct 16 AYHTLTAEEAAEFIGTSLTEGLTQDEFVHRLKTVGENTLGDDTKIDYKAMVLHQVCNAMI 75
Query 79 RILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIIL 114
+LL + ++SF A+ + GG+ ++V + +LI L
Sbjct 76 MVLLISMIISF--AMHDWITGGVISFVIAVNVLIGL 109
> sce:YGL167C PMR1, BSD1, LDB1, SSC1; High affinity Ca2+/Mn2+
P-type ATPase required for Ca2+ and Mn2+ transport into Golgi;
involved in Ca2+ dependent protein sorting and processing;
mutations in human homolog ATP2C1 cause acantholytic skin
condition Hailey-Hailey disease (EC:3.6.3.8); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=950
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 43/76 (56%), Gaps = 2/76 (2%)
Query 22 TCTVQEVLEYHETSLESGLPAEVVAE-RLERFGPNELQHEEGKSLFQLILEQF-QDLLVR 79
T +V E LE +T GL + A R +GPNE+ E+ +SLF+ L F +D ++
Sbjct 40 TLSVDEALEKLDTDKNGGLRSSNEANNRRSLYGPNEITVEDDESLFKKFLSNFIEDRMIL 99
Query 80 ILLAAAVVSFVLAVVE 95
+L+ +AVVS + ++
Sbjct 100 LLIGSAVVSLFMGNID 115
> cel:ZK256.1 pmr-1; PMR-type Golgi ATPase family member (pmr-1);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=978
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 33/93 (35%), Positives = 46/93 (49%), Gaps = 14/93 (15%)
Query 2 LMVATSPSEGRCRSLQEAPHT---CTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQ 58
+M+ T SE Q A H CT Q T+LE GL R + G NE
Sbjct 77 IMIETLTSE------QAASHEVVPCTHQ-----LRTNLEEGLTTAEATRRRQYHGYNEFD 125
Query 59 HEEGKSLFQLILEQFQDLLVRILLAAAVVSFVL 91
E + +++ LEQFQ+ L+ +LLA+A VS V+
Sbjct 126 VGEEEPIYKKYLEQFQNPLILLLLASAFVSIVM 158
> sce:YDR039C ENA2; Ena2p (EC:3.6.3.7 3.6.3.12); K01536 Na+-exporting
ATPase [EC:3.6.3.7]
Length=1091
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 49/96 (51%), Gaps = 2/96 (2%)
Query 19 APHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLV 78
A HT T +E E+ TSL GL + RL+ G N L + ++L Q + ++
Sbjct 16 AYHTLTTEEAAEFIGTSLTEGLTQDESLRRLKAVGENTLGDDTKIDYKAMVLHQVCNAMI 75
Query 79 RILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIIL 114
+L+ + +SF AV + GG+ ++V + +LI L
Sbjct 76 MVLVISMAISF--AVRDWITGGVISFVIAVNVLIGL 109
> sce:YDR038C ENA5; Ena5p (EC:3.6.3.7)
Length=1091
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 49/96 (51%), Gaps = 2/96 (2%)
Query 19 APHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLV 78
A HT T +E E+ TSL GL + RL+ G N L + ++L Q + ++
Sbjct 16 AYHTLTTEEAAEFIGTSLTEGLTQDESLRRLKAVGENTLGDDTKIDYKAMVLHQVCNAMI 75
Query 79 RILLAAAVVSFVLAVVEGGEGGLAAYVEPLVILIIL 114
+L+ + +SF AV + GG+ ++V + +LI L
Sbjct 76 MVLVISMAISF--AVRDWITGGVISFVIAVNVLIGL 109
> xla:399450 atp4a; ATPase, H+/K+ exchanging, alpha polypeptide
(EC:3.6.3.10); K01542 H+/K+-exchanging ATPase alpha polypeptide
[EC:3.6.3.10]
Length=1031
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 21 HTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRI 80
H TV+E+ + + TS+ GL + AE + R GPNEL+ +G + Q L +
Sbjct 51 HEITVEELEQKYTTSVSKGLKSAFAAEVILRDGPNELKPPKGTPEYIKFARQLAGGLQCL 110
Query 81 LLAAAVVSFVLAVVEGGEGGLAA 103
+ AAV+ + +E +G L +
Sbjct 111 MWVAAVICLIAFGIEESQGDLTS 133
> xla:733327 hypothetical protein LOC733327
Length=1031
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 21 HTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRI 80
H TV+E+ + + TS+ GL + AE + R GPNEL+ +G + Q L +
Sbjct 51 HEITVEELEQKYTTSVSKGLKSAFAAEVILRDGPNELKPPKGTPEYIKFARQLAGGLQCL 110
Query 81 LLAAAVVSFVLAVVEGGEGGLAA 103
+ AAV+ + +E +G L +
Sbjct 111 MWVAAVICLIAFGIEESQGDLTS 133
> cel:R05C11.3 ATPase; hypothetical protein; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1158
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 29/85 (34%), Positives = 39/85 (45%), Gaps = 18/85 (21%)
Query 10 EGRCRSLQEAPHTCTVQEVLEYHETSLESGLPAEV--VAERLERFGPNELQHEEGKSLFQ 67
EG CR L+ P +GLP + + R FG NE+ KS F+
Sbjct 37 EGLCRKLKTDP----------------INGLPNDTKELEHRRTAFGKNEIPPAPSKSFFR 80
Query 68 LILEQFQDLLVRILLAAAVVSFVLA 92
L E QD+ + ILL AA+VS L+
Sbjct 81 LAWEALQDITLVILLVAALVSLGLS 105
> ath:AT4G29900 ACA10; ACA10 (AUTOINHIBITED CA(2+)-ATPASE 10);
calcium-transporting ATPase/ calmodulin binding; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1069
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 3/71 (4%)
Query 33 ETSLESGLPAEV--VAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAAAVVSFV 90
+T+LE G+ + + +R FG N ++G+S ++ + E QDL + IL+ AAV S
Sbjct 147 KTNLEKGIHGDDDDILKRKSAFGSNTYPQKKGRSFWRFVWEASQDLTLIILIVAAVASLA 206
Query 91 LAV-VEGGEGG 100
L + EG E G
Sbjct 207 LGIKTEGIEKG 217
> cel:Y67D8C.10 mca-3; Membrane Calcium ATPase family member (mca-3);
K05850 Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1137
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 35/67 (52%), Gaps = 6/67 (8%)
Query 30 EYHETSLESGLPAEVVAERLER----FGPNELQHEEGKSLFQLILEQFQDLLVRILLAAA 85
E +T +GLP E LER FG NE+ K QL+ E QD+ + ILL +A
Sbjct 38 ERLKTDPNNGLPNN--EEELERRRNVFGANEIPPHPPKCFLQLVWEALQDVTLVILLVSA 95
Query 86 VVSFVLA 92
+VS L+
Sbjct 96 IVSLALS 102
> sce:YGL006W PMC1; Pmc1p (EC:3.6.3.8); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1173
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 11/80 (13%)
Query 48 RLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAAAVVSFVLAVV-----------EG 96
R + +G N L KS QL+ F D +++L AAVVSFVL + EG
Sbjct 87 RYKNYGDNSLPERIPKSFLQLVWAAFNDKTMQLLTVAAVVSFVLGLYELWMQPPQYDPEG 146
Query 97 GEGGLAAYVEPLVILIILFL 116
+ ++E + I+I +F+
Sbjct 147 NKIKQVDWIEGVAIMIAVFV 166
> mmu:11944 Atp4a; ATPase, H+/K+ exchanging, gastric, alpha polypeptide
(EC:3.6.3.10); K01542 H+/K+-exchanging ATPase alpha
polypeptide [EC:3.6.3.10]
Length=1025
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 21 HTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRI 80
H +V E+ + ++TS GL A + AE L R GPN L+ G + Q L +
Sbjct 54 HQLSVSELEQKYQTSATKGLKASLAAELLLRDGPNALRPPRGTPEYVKFARQLAGGLQCL 113
Query 81 LLAAAVVSFVLAVVEGGEGGLAA 103
+ AA + + ++ EG L
Sbjct 114 MWVAAAICLIAFAIQASEGDLTT 136
> cel:C02E7.1 hypothetical protein; K01539 sodium/potassium-transporting
ATPase subunit alpha [EC:3.6.3.9]
Length=1050
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 29 LEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAAAVVS 88
L+ + + GL +V AERLE G N L + S ++L L QF++LL ++ AA +S
Sbjct 56 LDIVDPKVSRGLSKQVAAERLESDGRNALSPPKVISNWKLFLRQFKNLLWILMFGAAALS 115
Query 89 FV 90
V
Sbjct 116 LV 117
> ath:AT3G42640 AHA8; AHA8 (Arabidopsis H(+)-ATPase 8); ATPase;
K01535 H+-transporting ATPase [EC:3.6.3.6]
Length=948
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 58/101 (57%), Gaps = 5/101 (4%)
Query 25 VQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAA 84
V+EV E + S E GL ++ A+RLE FG N+L+ E+ ++ F L + L ++ +A
Sbjct 22 VEEVFEQLKCSKE-GLSSDEGAKRLEIFGANKLE-EKSENKFLKFLGFMWNPLSWVMESA 79
Query 85 AVVSFVLAVVEGGEGGLAAYVEPLVILIILFLNAAVXVWQE 125
A+++ VLA G G + + + I+++L +N+ + +E
Sbjct 80 AIMAIVLA---NGGGKAPDWQDFIGIMVLLIINSTISFIEE 117
> ath:AT3G22910 calcium-transporting ATPase, plasma membrane-type,
putative / Ca(2+)-ATPase, putative (ACA13); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1017
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 45 VAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAAAVVSFVLAVVEGG 97
+ R FG N + K LF ++E F+DL + ILL A +S + E G
Sbjct 128 IQRRRSTFGSNTYTRQPSKGLFHFVVEAFKDLTILILLGCATLSLGFGIKEHG 180
> hsa:495 ATP4A, ATP6A; ATPase, H+/K+ exchanging, alpha polypeptide
(EC:3.6.3.10); K01542 H+/K+-exchanging ATPase alpha polypeptide
[EC:3.6.3.10]
Length=1035
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 21 HTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRI 80
H +V E+ + ++TS GL A + AE L R GPN L+ G + Q L +
Sbjct 55 HQLSVAELEQKYQTSATKGLSASLAAELLLRDGPNALRPPRGTPEYVKFARQLAGGLQCL 114
Query 81 LLAAAVVSFVLAVVEGGEGGLAA 103
+ AA + + ++ EG L
Sbjct 115 MWVAAAICLIAFAIQASEGDLTT 137
> dre:768304 atp2b4, MGC152801, zgc:152801; ATPase, Ca++ transporting,
plasma membrane 4 (EC:3.6.3.8)
Length=316
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 45/94 (47%), Gaps = 14/94 (14%)
Query 10 EGRCRSLQEAPHTCTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLI 69
+G CR L+ +P SG PA++ +R FG N + ++ K+ QL+
Sbjct 53 QGICRRLKTSPIEGL-------------SGNPADI-EKRHTSFGKNFIPPKKPKTFLQLV 98
Query 70 LEQFQDLLVRILLAAAVVSFVLAVVEGGEGGLAA 103
E QD+ + IL AA++S L+ EG AA
Sbjct 99 WEALQDVTLIILEVAAIISLALSFYHPPEGDNAA 132
> dre:566794 atp2c1; ATPase, Ca++ transporting, type 2C, member
1; K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=925
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 0/67 (0%)
Query 25 VQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAA 84
V EV + L+ GL E V R G NE E + L++ L QF+D L+ +LL++
Sbjct 38 VNEVACALQADLQFGLNREEVERRRTYHGWNEFDISEDEPLWKKYLSQFKDPLILLLLSS 97
Query 85 AVVSFVL 91
AV+S ++
Sbjct 98 AVISVLM 104
> mmu:235574 Atp2c1, 1700121J11Rik, ATP2C1A, AW061228, BCPM, D930003G21Rik,
HHD, MGC58010, SPCA, pmr1; ATPase, Ca++-sequestering
(EC:3.6.3.8); K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=918
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 38/67 (56%), Gaps = 0/67 (0%)
Query 25 VQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAA 84
V EV + L++GL V+ R G NE E + L++ + QF++ L+ +LLA+
Sbjct 31 VSEVAGLLQADLQNGLNKSEVSHRRAFHGWNEFDISEDEPLWKKYISQFKNPLIMLLLAS 90
Query 85 AVVSFVL 91
AV+S ++
Sbjct 91 AVISILM 97
> mmu:69047 Atp2c2, 1810010G06Rik, KIAA0703, Spca2, mKIAA0703;
ATPase, Ca++ transporting, type 2C, member 2 (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=944
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 38/66 (57%), Gaps = 0/66 (0%)
Query 23 CTVQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILL 82
C+ +E+ L+SGL VA+R G NE + + +++ L+QF++ L+ +LL
Sbjct 58 CSREELARAFHVDLDSGLSEFAVAQRRLVHGWNEFVTDNAEPVWKKYLDQFRNPLILLLL 117
Query 83 AAAVVS 88
++VVS
Sbjct 118 GSSVVS 123
> dre:561190 atp2b1b; ATPase, Ca++ transporting, plasma membrane
1b
Length=1240
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 38 SGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAAAVVSFVLA 92
SG P ++ A R E FG N + ++ K+ QL+ E QD+ + IL AA++S L+
Sbjct 66 SGHPDDI-ARRKEEFGKNFIPPKKPKTFLQLVWEALQDVTLIILEVAAIISLGLS 119
> hsa:27032 ATP2C1, ATP2C1A, BCPM, HHD, KIAA1347, PMR1, SPCA1,
hSPCA1; ATPase, Ca++ transporting, type 2C, member 1 (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=888
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 38/67 (56%), Gaps = 0/67 (0%)
Query 25 VQEVLEYHETSLESGLPAEVVAERLERFGPNELQHEEGKSLFQLILEQFQDLLVRILLAA 84
V EV + L++GL V+ R G NE E + L++ + QF++ L+ +LLA+
Sbjct 31 VSEVASILQADLQNGLNKCEVSHRRAFHGWNEFDISEDEPLWKKYISQFKNPLIMLLLAS 90
Query 85 AVVSFVL 91
AV+S ++
Sbjct 91 AVISVLM 97
Lambda K H
0.320 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2049573556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40