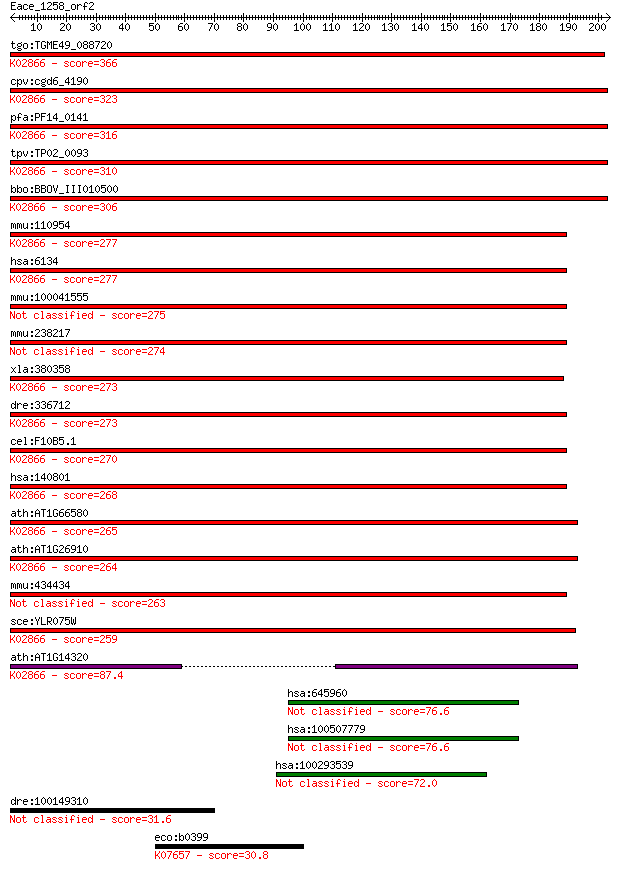
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1258_orf2
Length=203
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_088720 60S ribosomal protein L10, putative ; K02866... 366 3e-101
cpv:cgd6_4190 60S ribosomal protein L10, alpha/beta hammerhead... 323 3e-88
pfa:PF14_0141 60S ribosomal protein L10, putative; K02866 larg... 316 4e-86
tpv:TP02_0093 60S ribosomal protein L10; K02866 large subunit ... 310 3e-84
bbo:BBOV_III010500 17.m07908; ribosomal protein L10; K02866 la... 306 3e-83
mmu:110954 Rpl10, D0HXS648, DXHXS648, DXHXS648E, MGC107630, QM... 277 2e-74
hsa:6134 RPL10, DKFZp686J1851, DXS648, DXS648E, FLJ23544, FLJ2... 277 2e-74
mmu:100041555 Gm3405; predicted gene 3405 275 8e-74
mmu:238217 Rpl10l, EG238217; ribosomal protein L10-like 274 1e-73
xla:380358 rpl10, MGC53488; ribosomal protein L10 (EC:3.6.5.3)... 273 4e-73
dre:336712 rpl10, QM, fa93d03, rpl-10, wu:fa93d03, zgc:56154; ... 273 4e-73
cel:F10B5.1 rpl-10; Ribosomal Protein, Large subunit family me... 270 3e-72
hsa:140801 RPL10L, FLJ27353, RPL10_5_1358; ribosomal protein L... 268 7e-72
ath:AT1G66580 60S ribosomal protein L10 (RPL10C); K02866 large... 265 1e-70
ath:AT1G26910 60S ribosomal protein L10 (RPL10B); K02866 large... 264 1e-70
mmu:434434 Gm5621, EG434434; predicted gene 5621 263 4e-70
sce:YLR075W RPL10, GRC5, QSR1; L10; K02866 large subunit ribos... 259 6e-69
ath:AT1G14320 SAC52; SAC52 (SUPPRESSOR OF ACAULIS 52); structu... 87.4 3e-17
hsa:645960 similar to hCG1644362 76.6 5e-14
hsa:100507779 hypothetical protein LOC100507779 76.6 5e-14
hsa:100293539 hypothetical protein LOC100293539 72.0 1e-12
dre:100149310 syt6a, si:ch73-306c3.1; synaptotagmin VIa 31.6 1.9
eco:b0399 phoB, ECK0393, JW0389, phoRc; DNA-binding response r... 30.8 3.7
> tgo:TGME49_088720 60S ribosomal protein L10, putative ; K02866
large subunit ribosomal protein L10e
Length=221
Score = 366 bits (939), Expect = 3e-101, Method: Compositional matrix adjust.
Identities = 174/201 (86%), Positives = 185/201 (92%), Gaps = 0/201 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RIYD GRK+ADVDEFPGV HLVSDEYEQISSEALEAARVCANKYMIK+
Sbjct 20 SRFCRGVPDAKIRIYDAGRKRADVDEFPGVAHLVSDEYEQISSEALEAARVCANKYMIKN 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKDNFHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP GLVARV+IGQILMSIR
Sbjct 80 CGKDNFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPMGLVARVDIGQILMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
T+ES V A ALRRAAFKFPGRQKVF SNKWGFT FT+ EYKKWQAEGRIVSDGV AKW
Sbjct 140 TRESNVATAVTALRRAAFKFPGRQKVFVSNKWGFTKFTKVEYKKWQAEGRIVSDGVGAKW 199
Query 181 LAKKGPLSDTFPLAKDIHLPV 201
++ KGPL++TFP A DI LPV
Sbjct 200 ISTKGPLANTFPSAADITLPV 220
> cpv:cgd6_4190 60S ribosomal protein L10, alpha/beta hammerhead
; K02866 large subunit ribosomal protein L10e
Length=222
Score = 323 bits (828), Expect = 3e-88, Method: Compositional matrix adjust.
Identities = 151/202 (74%), Positives = 173/202 (85%), Gaps = 0/202 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPD K+RIYDVGRKKA+ D FP V+HL+SDEYEQISSEALEAAR+ ANKYMIK
Sbjct 20 SRFCRGVPDPKIRIYDVGRKKAECDAFPAVIHLISDEYEQISSEALEAARISANKYMIKF 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKDNFHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGA+GKPTG ARVNIGQ+LMSIR
Sbjct 80 CGKDNFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAYGKPTGTAARVNIGQVLMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
KE A +ALRRA +KFPGRQKV S+KWGFT FT+EEY K+QAEGRIV DGV K+
Sbjct 140 CKEDKTQTAVMALRRAKYKFPGRQKVVVSDKWGFTKFTKEEYLKYQAEGRIVPDGVNCKY 199
Query 181 LAKKGPLSDTFPLAKDIHLPVE 202
++ +G LS FP A +I++P++
Sbjct 200 ISCRGSLSRIFPEAANINIPLD 221
> pfa:PF14_0141 60S ribosomal protein L10, putative; K02866 large
subunit ribosomal protein L10e
Length=219
Score = 316 bits (809), Expect = 4e-86, Method: Compositional matrix adjust.
Identities = 148/202 (73%), Positives = 178/202 (88%), Gaps = 3/202 (1%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SR+CRGVPD K+RIYD+GRKKADV+EF GVVHLVS EYEQISSEALEAAR+ ANKYMI +
Sbjct 20 SRYCRGVPDPKIRIYDMGRKKADVNEFSGVVHLVSYEYEQISSEALEAARISANKYMITN 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKDNFHLR+R+HPFHVLRINKMLSCAGADRLQTGMRGAFGKP G+VARV+IGQ+L+SIR
Sbjct 80 CGKDNFHLRVRIHPFHVLRINKMLSCAGADRLQTGMRGAFGKPNGVVARVDIGQVLLSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TKE+ V+ A ALRRA +KFPGRQKVF SNKWGFTPF+++EY++++ +GRI+SDGV K+
Sbjct 140 TKENFVSKACEALRRAKYKFPGRQKVFVSNKWGFTPFSKDEYQQYKKKGRIISDGVSCKF 199
Query 181 LAKKGPLSDTFPLAKDIHLPVE 202
+ +KGPL + KDI+ +E
Sbjct 200 IREKGPLDKIY---KDINTVLE 218
> tpv:TP02_0093 60S ribosomal protein L10; K02866 large subunit
ribosomal protein L10e
Length=222
Score = 310 bits (793), Expect = 3e-84, Method: Compositional matrix adjust.
Identities = 144/202 (71%), Positives = 172/202 (85%), Gaps = 0/202 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPD K+RIYD+G K ADVD+FP VH+VS EYEQISSEALEAAR+CANK+M+K
Sbjct 20 SRFCRGVPDPKIRIYDMGLKGADVDDFPCAVHIVSGEYEQISSEALEAARICANKFMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
GK++FH+R+RVHPFHVLRINKMLSCAGADRLQTGMR AFGKPTG+VARVNIGQ+LMSIR
Sbjct 80 GGKESFHIRVRVHPFHVLRINKMLSCAGADRLQTGMRRAFGKPTGVVARVNIGQVLMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
T+E V A ALRRA +KFPGRQK+F SNKWGFTPFT+EEY K+Q+EGR+ + GV KW
Sbjct 140 TREHLVPKAIEALRRAKYKFPGRQKIFVSNKWGFTPFTKEEYLKYQSEGRLENKGVHVKW 199
Query 181 LAKKGPLSDTFPLAKDIHLPVE 202
++ GPL+ FP A + +P++
Sbjct 200 ISSHGPLTKIFPNADSVSVPLD 221
> bbo:BBOV_III010500 17.m07908; ribosomal protein L10; K02866
large subunit ribosomal protein L10e
Length=227
Score = 306 bits (785), Expect = 3e-83, Method: Compositional matrix adjust.
Identities = 141/202 (69%), Positives = 173/202 (85%), Gaps = 0/202 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RIYD+G K ADVD+FP VHLVS EYEQISSEALEAAR+CANK+M+K
Sbjct 20 SRFCRGVPDAKIRIYDMGLKGADVDDFPYAVHLVSGEYEQISSEALEAARICANKFMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
GK++FH+R+RVHPFHVLRINKMLSCAGADRLQTGMR AFGKP G+VARV+IGQIL+S+R
Sbjct 80 GGKESFHIRIRVHPFHVLRINKMLSCAGADRLQTGMRRAFGKPNGVVARVDIGQILISVR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
T+E+ V A ALRRA +KFPGRQK+F SN WGFT FTRE+Y K+Q+EGR+ + GV KW
Sbjct 140 TREALVPKAVEALRRAKYKFPGRQKIFISNNWGFTNFTREDYLKYQSEGRLENKGVHVKW 199
Query 181 LAKKGPLSDTFPLAKDIHLPVE 202
++ GPL+ FP +++H+P++
Sbjct 200 ISSHGPLTKVFPQLENVHVPLD 221
> mmu:110954 Rpl10, D0HXS648, DXHXS648, DXHXS648E, MGC107630,
QM, QM16; ribosomal protein 10; K02866 large subunit ribosomal
protein L10e
Length=214
Score = 277 bits (708), Expect = 2e-74, Method: Compositional matrix adjust.
Identities = 130/188 (69%), Positives = 153/188 (81%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDAKIRIFDLGRKKAKVDEFPLCGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARV+IGQ++MSIR
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQVIMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S KWGFT F +E++ AE R++ DG K+
Sbjct 140 TKLQNKEHVIEALRRAKFKFPGRQKIHISKKWGFTKFNADEFEDMVAEKRLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ +GPL
Sbjct 200 IPNRGPLD 207
> hsa:6134 RPL10, DKFZp686J1851, DXS648, DXS648E, FLJ23544, FLJ27072,
NOV, QM; ribosomal protein L10; K02866 large subunit
ribosomal protein L10e
Length=214
Score = 277 bits (708), Expect = 2e-74, Method: Compositional matrix adjust.
Identities = 130/188 (69%), Positives = 153/188 (81%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDAKIRIFDLGRKKAKVDEFPLCGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARV+IGQ++MSIR
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQVIMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S KWGFT F +E++ AE R++ DG K+
Sbjct 140 TKLQNKEHVIEALRRAKFKFPGRQKIHISKKWGFTKFNADEFEDMVAEKRLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ +GPL
Sbjct 200 IPNRGPLD 207
> mmu:100041555 Gm3405; predicted gene 3405
Length=214
Score = 275 bits (703), Expect = 8e-74, Method: Compositional matrix adjust.
Identities = 130/188 (69%), Positives = 152/188 (80%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDAKTRIFDLGRKKAKVDEFPLCGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARV+IGQ++MSIR
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQVIMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S KWGFT F +E++ AE R++ DG K+
Sbjct 140 TKLQNKEHVIEALRRAKFKFPGRQKIHISKKWGFTKFNADEFEDMVAEKRLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ +GPL
Sbjct 200 IPNRGPLD 207
> mmu:238217 Rpl10l, EG238217; ribosomal protein L10-like
Length=214
Score = 274 bits (701), Expect = 1e-73, Method: Compositional matrix adjust.
Identities = 129/188 (68%), Positives = 153/188 (81%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDAKIRIFDLGRKKAKVDEFPLCGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARV+IGQ++MSIR
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQVIMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S KWGFT F +E++ A R++ DG K+
Sbjct 140 TKLQNKEHVIEALRRAKFKFPGRQKIHISKKWGFTKFNADEFEDKVAAKRLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ ++GPL
Sbjct 200 IPERGPLD 207
> xla:380358 rpl10, MGC53488; ribosomal protein L10 (EC:3.6.5.3);
K02866 large subunit ribosomal protein L10e
Length=215
Score = 273 bits (697), Expect = 4e-73, Method: Compositional matrix adjust.
Identities = 128/187 (68%), Positives = 150/187 (80%), Gaps = 0/187 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPD K+RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDPKIRIFDLGRKKAKVDEFPLCGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARVNIGQ++MSIR
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVNIGQVIMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S K+GFT F + ++ AE R++ DG K+
Sbjct 140 TKTQNKEHVIEALRRAKFKFPGRQKIHISKKFGFTKFNADNFESMLAEKRLIPDGCGVKY 199
Query 181 LAKKGPL 187
+ GPL
Sbjct 200 IPNSGPL 206
> dre:336712 rpl10, QM, fa93d03, rpl-10, wu:fa93d03, zgc:56154;
ribosomal protein L10; K02866 large subunit ribosomal protein
L10e
Length=215
Score = 273 bits (697), Expect = 4e-73, Method: Compositional matrix adjust.
Identities = 127/188 (67%), Positives = 151/188 (80%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPD K+RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDPKIRIFDLGRKKAKVDEFPLCAHMVSDEYEQLSSEALEAARICANKYMVKT 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARV+IGQ++MS+R
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQVIMSVR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S K+GFT F ++ AE R++ DG K+
Sbjct 140 TKAQNKEHVIEALRRAKFKFPGRQKIHVSKKYGFTKFNTCDFDNMLAEKRLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ +GPLS
Sbjct 200 IPSRGPLS 207
> cel:F10B5.1 rpl-10; Ribosomal Protein, Large subunit family
member (rpl-10); K02866 large subunit ribosomal protein L10e
Length=214
Score = 270 bits (690), Expect = 3e-72, Method: Compositional matrix adjust.
Identities = 127/188 (67%), Positives = 154/188 (81%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RI+D+G K+A+VD FP VH++S+E E +SSEALEAAR+CANKYM+K+
Sbjct 20 SRFCRGVPDAKIRIFDLGNKRANVDTFPACVHMMSNEREHLSSEALEAARICANKYMVKN 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FHLR+R HPFHV RINKMLSCAGADRLQTGMRGA+GKP GLVARV+IG IL S+R
Sbjct 80 CGKDGFHLRVRKHPFHVTRINKMLSCAGADRLQTGMRGAYGKPQGLVARVDIGDILFSMR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
KE V A A RRA FKFPGRQ + +S KWGFT + RE+Y++ +AEGR+ SDGV +
Sbjct 140 IKEGNVKHAIEAFRRAKFKFPGRQIIVSSRKWGFTKWDREDYERMRAEGRLRSDGVGVQL 199
Query 181 LAKKGPLS 188
+ GPL+
Sbjct 200 QREHGPLT 207
> hsa:140801 RPL10L, FLJ27353, RPL10_5_1358; ribosomal protein
L10-like; K02866 large subunit ribosomal protein L10e
Length=214
Score = 268 bits (686), Expect = 7e-72, Method: Compositional matrix adjust.
Identities = 127/188 (67%), Positives = 151/188 (80%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RI+D+GRKKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDAKIRIFDLGRKKAKVDEFPLGGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CG+D FH+R+R+HPFHV+RINKMLSCAGADRLQTGMRGAFGKP G VARV+IGQ++MSIR
Sbjct 80 CGRDGFHMRVRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQVIMSIR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK ALRRA FKFPGRQK+ S KWGFT F +E++ A+ ++ DG K+
Sbjct 140 TKLQNEEHVIEALRRAKFKFPGRQKIHISKKWGFTKFNADEFEDMVAKKCLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ GPL
Sbjct 200 VPSHGPLD 207
> ath:AT1G66580 60S ribosomal protein L10 (RPL10C); K02866 large
subunit ribosomal protein L10e
Length=221
Score = 265 bits (676), Expect = 1e-70, Method: Compositional matrix adjust.
Identities = 131/192 (68%), Positives = 149/192 (77%), Gaps = 0/192 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SR+CRGVPD K+RIYDVG K+ VDEFP VHLVS E E +SSEALEAAR+ NKYM+K
Sbjct 20 SRYCRGVPDPKIRIYDVGMKRKGVDEFPFCVHLVSWEKENVSSEALEAARIACNKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
GKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK G ARV IGQ+L+S+R
Sbjct 80 AGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVLLSVR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
K++ A ALRRA FKFPGRQK+ S KWGFT F R EY K +A RIV DGV AK+
Sbjct 140 CKDNHGVHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRAEYTKLRAMKRIVPDGVNAKF 199
Query 181 LAKKGPLSDTFP 192
L+ GPL++ P
Sbjct 200 LSNHGPLANRQP 211
> ath:AT1G26910 60S ribosomal protein L10 (RPL10B); K02866 large
subunit ribosomal protein L10e
Length=221
Score = 264 bits (675), Expect = 1e-70, Method: Compositional matrix adjust.
Identities = 130/192 (67%), Positives = 150/192 (78%), Gaps = 0/192 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SR+CRGVPD K+RIYDVG K+ VDEFP VHLVS E E +SSEALEAAR+ NKYM+K
Sbjct 20 SRYCRGVPDPKIRIYDVGMKRKGVDEFPYCVHLVSWEKENVSSEALEAARIACNKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
GKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK G ARV IGQ+L+S+R
Sbjct 80 AGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVLLSVR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
K++ + A ALRRA FKFPGRQK+ S KWGFT F R +Y K + E RIV DGV AK+
Sbjct 140 CKDAHGHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRADYTKLRQEKRIVPDGVNAKF 199
Query 181 LAKKGPLSDTFP 192
L+ GPL++ P
Sbjct 200 LSCHGPLANRQP 211
> mmu:434434 Gm5621, EG434434; predicted gene 5621
Length=214
Score = 263 bits (671), Expect = 4e-70, Method: Compositional matrix adjust.
Identities = 124/188 (65%), Positives = 148/188 (78%), Gaps = 0/188 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SRFCRGVPDAK+RI+D+G KKA VDEFP H+VSDEYEQ+SSEALEAAR+CANKYM+K
Sbjct 20 SRFCRGVPDAKIRIFDLGWKKAKVDEFPLCGHMVSDEYEQLSSEALEAARICANKYMVKS 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
CGKD FH+R+R+HPFHV+RINKMLSCAGA+RLQTGMRGAFGKP G VARV+IGQ++ SI
Sbjct 80 CGKDGFHIRVRLHPFHVIRINKMLSCAGANRLQTGMRGAFGKPQGTVARVHIGQVITSIX 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
K ALRRA FKFPG QK+ S KWGFT F +E++ AE R++ DG K+
Sbjct 140 AKLQNKEHVIEALRRAKFKFPGHQKIHISKKWGFTKFNADEFEDMVAEKRLIPDGCGVKY 199
Query 181 LAKKGPLS 188
+ +GPL
Sbjct 200 IPNRGPLD 207
> sce:YLR075W RPL10, GRC5, QSR1; L10; K02866 large subunit ribosomal
protein L10e
Length=221
Score = 259 bits (661), Expect = 6e-69, Method: Compositional matrix adjust.
Identities = 123/191 (64%), Positives = 148/191 (77%), Gaps = 0/191 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMIKH 60
SR+ R VPD+K+RIYD+G+KKA VDEFP VHLVS+E EQ+SSEALEAAR+CANKYM
Sbjct 20 SRYNRAVPDSKIRIYDLGKKKATVDEFPLCVHLVSNELEQLSSEALEAARICANKYMTTV 79
Query 61 CGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPTGLVARVNIGQILMSIR 120
G+D FHLR+RVHPFHVLRINKMLSCAGADRLQ GMRGA+GKP GL ARV+IGQI+ S+R
Sbjct 80 SGRDAFHLRVRVHPFHVLRINKMLSCAGADRLQQGMRGAWGKPHGLAARVDIGQIIFSVR 139
Query 121 TKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGRIVSDGVCAKW 180
TK+S ++ LRRA +KFPG+QK+ S KWGFT R EY K + G + DG K+
Sbjct 140 TKDSNKDVVVEGLRRARYKFPGQQKIILSKKWGFTNLDRPEYLKKREAGEVKDDGAFVKF 199
Query 181 LAKKGPLSDTF 191
L+KKG L +
Sbjct 200 LSKKGSLENNI 210
> ath:AT1G14320 SAC52; SAC52 (SUPPRESSOR OF ACAULIS 52); structural
constituent of ribosome; K02866 large subunit ribosomal
protein L10e
Length=163
Score = 87.4 bits (215), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 41/58 (70%), Positives = 47/58 (81%), Gaps = 0/58 (0%)
Query 1 SRFCRGVPDAKLRIYDVGRKKADVDEFPGVVHLVSDEYEQISSEALEAARVCANKYMI 58
SR+CRGVPD K+RIYDVG K+ VDEFP VHLVS E E +SSEALEAAR+ NKYM+
Sbjct 20 SRYCRGVPDPKIRIYDVGMKRKGVDEFPFCVHLVSWEKENVSSEALEAARIACNKYMV 77
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/82 (50%), Positives = 56/82 (68%), Gaps = 0/82 (0%)
Query 111 NIGQILMSIRTKESTVNIASLALRRAAFKFPGRQKVFTSNKWGFTPFTREEYKKWQAEGR 170
N +L+S+R K++ + A ALRRA FKFPGRQK+ S KWGFT F R ++ K + E R
Sbjct 73 NKYMVLLSVRCKDAHGHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRADFTKLRQEKR 132
Query 171 IVSDGVCAKWLAKKGPLSDTFP 192
+V DGV AK+L+ GPL++ P
Sbjct 133 VVPDGVNAKFLSCHGPLANRQP 154
> hsa:645960 similar to hCG1644362
Length=226
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 38/78 (48%), Positives = 51/78 (65%), Gaps = 0/78 (0%)
Query 95 GMRGAFGKPTGLVARVNIGQILMSIRTKESTVNIASLALRRAAFKFPGRQKVFTSNKWGF 154
+RGAF KP G VARV+IGQ++MSI TK AL +A FKFPGRQ + S KW F
Sbjct 27 SIRGAFEKPQGAVARVHIGQVIMSICTKLQNKEHVIEALCKANFKFPGRQNIHFSEKWDF 86
Query 155 TPFTREEYKKWQAEGRIV 172
T F+ +E++ AE +++
Sbjct 87 TKFSVDEFEDMMAEKQLI 104
> hsa:100507779 hypothetical protein LOC100507779
Length=226
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 38/78 (48%), Positives = 51/78 (65%), Gaps = 0/78 (0%)
Query 95 GMRGAFGKPTGLVARVNIGQILMSIRTKESTVNIASLALRRAAFKFPGRQKVFTSNKWGF 154
+RGAF KP G VARV+IGQ++MSI TK AL +A FKFPGRQ + S KW F
Sbjct 27 SIRGAFEKPQGAVARVHIGQVIMSICTKLQNKEHVIEALCKANFKFPGRQNIHFSEKWDF 86
Query 155 TPFTREEYKKWQAEGRIV 172
T F+ +E++ AE +++
Sbjct 87 TKFSVDEFEDMMAEKQLI 104
> hsa:100293539 hypothetical protein LOC100293539
Length=353
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 40/71 (56%), Positives = 47/71 (66%), Gaps = 1/71 (1%)
Query 91 RLQTGMRGAFGKPTGLVARVNIGQILMSIRTKESTVNIASLALRRAAFKFPGRQKVFTSN 150
RL+TG++GAFGKP G VARV+IGQ+ SI TK A RA FKFPG QK+ S
Sbjct 284 RLETGIQGAFGKPQGTVARVHIGQV-KSICTKLQNKEHVIEAPCRAKFKFPGHQKIHISK 342
Query 151 KWGFTPFTREE 161
KWGFT F +E
Sbjct 343 KWGFTKFNVDE 353
> dre:100149310 syt6a, si:ch73-306c3.1; synaptotagmin VIa
Length=555
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 38/71 (53%), Gaps = 7/71 (9%)
Query 1 SRFCRGVP-DAKLRIYDVGRKKADVDEFPGVVHLVSDE-YEQISSEALEAARVCANKYMI 58
S F R +P ++ D+G DVDE P + + E Y+Q+++E E+A N
Sbjct 200 SSFKRHLPRQMQVSSLDLG-DDYDVDEQPTSIGRIKPELYKQMTTENDESA----NSKGG 254
Query 59 KHCGKDNFHLR 69
K+CGK NF LR
Sbjct 255 KNCGKINFSLR 265
> eco:b0399 phoB, ECK0393, JW0389, phoRc; DNA-binding response
regulator in two-component regulatory system with PhoR (or
CreC); K07657 two-component system, OmpR family, phosphate regulon
response regulator PhoB
Length=229
Score = 30.8 bits (68), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 14/50 (28%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 50 RVCANKYMIKHCGKDNFHLRMRVHPFHVLRINKMLSCAGADRLQTGMRGA 99
RV + + ++ H N ++ R H+ R+ K L G DR+ +RG
Sbjct 172 RVYSREQLLNHVWGTNVYVEDRTVDVHIRRLRKALEPGGHDRMVQTVRGT 221
Lambda K H
0.324 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6189150336
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40