bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1311_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
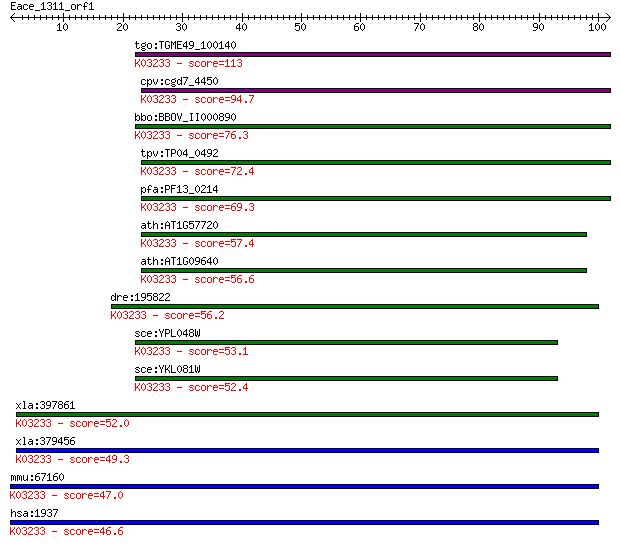
tgo:TGME49_100140 elongation factor 1-gamma, putative (EC:2.5.... 113 1e-25
cpv:cgd7_4450 elongation factor EF1-gamma (glutathione S-trans... 94.7 7e-20
bbo:BBOV_II000890 18.m06063; hypothetical protein; K03233 elon... 76.3 2e-14
tpv:TP04_0492 elongation factor 1 gamma (EC:3.6.1.48); K03233 ... 72.4 3e-13
pfa:PF13_0214 elongation factor 1-gamma, putative; K03233 elon... 69.3 3e-12
ath:AT1G57720 elongation factor 1B-gamma, putative / eEF-1B ga... 57.4 1e-08
ath:AT1G09640 elongation factor 1B-gamma, putative / eEF-1B ga... 56.6 2e-08
dre:195822 eef1g, db03h10, fk76a09, mg:db03h10, wu:fk76a09; eu... 56.2 3e-08
sce:YPL048W CAM1, TEF3; CPBP; K03233 elongation factor 1-gamma 53.1 2e-07
sce:YKL081W TEF4, EFC1; Tef4p; K03233 elongation factor 1-gamma 52.4 4e-07
xla:397861 eef1g-b, ef1g, gig35; eukaryotic translation elonga... 52.0 5e-07
xla:379456 eef1g-a, MGC64329, eef1g, ef1g, gig35; eukaryotic t... 49.3 3e-06
mmu:67160 Eef1g, 2610301D06Rik, AA407312, EF1G, MGC103354, MGC... 47.0 2e-05
hsa:1937 EEF1G, EF1G, GIG35; eukaryotic translation elongation... 46.6 2e-05
> tgo:TGME49_100140 elongation factor 1-gamma, putative (EC:2.5.1.18);
K03233 elongation factor 1-gamma
Length=394
Score = 113 bits (283), Expect = 1e-25, Method: Composition-based stats.
Identities = 50/81 (61%), Positives = 66/81 (81%), Gaps = 1/81 (1%)
Query 22 ESSLDMNEWKKVYSNTQDLRGVAMKWLWDHVDEH-CSFFLMEYEKIEGECTVTYLTSNQL 80
E ++D+NEWK+VYSNT+DL G AMKW W+H+D S + M+Y+K+EGECTV ++TSNQL
Sbjct 243 EPTMDLNEWKRVYSNTKDLYGTAMKWFWEHLDAAGYSLWYMKYQKLEGECTVAFVTSNQL 302
Query 81 GGFLQRVDNSMRKDSFAVIDV 101
GGFLQR+D + RK SF V+DV
Sbjct 303 GGFLQRIDPAFRKYSFGVVDV 323
> cpv:cgd7_4450 elongation factor EF1-gamma (glutathione S-transferase
family) ; K03233 elongation factor 1-gamma
Length=382
Score = 94.7 bits (234), Expect = 7e-20, Method: Composition-based stats.
Identities = 42/80 (52%), Positives = 60/80 (75%), Gaps = 1/80 (1%)
Query 23 SSLDMNEWKKVYSNTQDLRGVAMKWLWDHVDEH-CSFFLMEYEKIEGECTVTYLTSNQLG 81
+S+ M+EWK+VYSNT+DL+GVAM WL+ + D + SF+ M+Y K+ E V + SN +G
Sbjct 232 TSMSMDEWKRVYSNTKDLKGVAMPWLYQNFDANGYSFYYMKYNKLPDELDVAFRASNMVG 291
Query 82 GFLQRVDNSMRKDSFAVIDV 101
GFLQR+DN+ RK SF VI++
Sbjct 292 GFLQRLDNNFRKYSFGVINI 311
> bbo:BBOV_II000890 18.m06063; hypothetical protein; K03233 elongation
factor 1-gamma
Length=360
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/82 (45%), Positives = 51/82 (62%), Gaps = 2/82 (2%)
Query 22 ESSLDMNEWKKVYSNTQ-DLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLTSNQ 79
ES ++ WKK YSN + DL M WLWD+ D ++ +F+ M+Y K++ EC TSN
Sbjct 208 ESGFVIDTWKKTYSNCKGDLEKEVMPWLWDNFDSDNWNFYFMKYNKLDDECKSEITTSNM 267
Query 80 LGGFLQRVDNSMRKDSFAVIDV 101
L GFLQR + R+ SF VI+V
Sbjct 268 LSGFLQRFEPDFRRISFGVINV 289
> tpv:TP04_0492 elongation factor 1 gamma (EC:3.6.1.48); K03233
elongation factor 1-gamma
Length=388
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 48/81 (59%), Gaps = 2/81 (2%)
Query 23 SSLDMNEWKKVYSNTQ-DLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLTSNQL 80
+S +++WKK YSN + DL M WLW+ D E S + M Y K+E EC TSN L
Sbjct 237 TSFSLDQWKKTYSNCKGDLYKEVMPWLWEKFDPEGWSLYYMVYNKLEDECKSEVFTSNML 296
Query 81 GGFLQRVDNSMRKDSFAVIDV 101
GFLQR +N R SF V++V
Sbjct 297 SGFLQRFENEFRHYSFGVLNV 317
> pfa:PF13_0214 elongation factor 1-gamma, putative; K03233 elongation
factor 1-gamma
Length=411
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 33/80 (41%), Positives = 51/80 (63%), Gaps = 1/80 (1%)
Query 23 SSLDMNEWKKVYSNTQDLRGVAMKWLWDHVDEH-CSFFLMEYEKIEGECTVTYLTSNQLG 81
S+ ++EWK +SN +DL AM W D + S + M+Y+K+E EC ++++ N G
Sbjct 261 SNFSLDEWKYKFSNEKDLLNNAMPHFWKTYDPNGFSLYYMKYDKLEDECQISFVACNMAG 320
Query 82 GFLQRVDNSMRKDSFAVIDV 101
GFLQR++N+ K SFAV+ V
Sbjct 321 GFLQRLENNFSKYSFAVVTV 340
> ath:AT1G57720 elongation factor 1B-gamma, putative / eEF-1B
gamma, putative; K03233 elongation factor 1-gamma
Length=413
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 33/77 (42%), Positives = 51/77 (66%), Gaps = 4/77 (5%)
Query 23 SSLDMNEWKKVYSNTQ-DLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLTSNQL 80
S + +++WK++YSNT+ + R VA+K WD D E S + +Y K E V+++T N++
Sbjct 263 SPMVLDDWKRLYSNTKSNFREVAIKGFWDMYDPEGYSLWFCDY-KYNDENMVSFVTLNKV 321
Query 81 GGFLQRVDNSMRKDSFA 97
GGFLQR+D + RK SF
Sbjct 322 GGFLQRMDLA-RKYSFG 337
> ath:AT1G09640 elongation factor 1B-gamma, putative / eEF-1B
gamma, putative; K03233 elongation factor 1-gamma
Length=414
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 33/77 (42%), Positives = 51/77 (66%), Gaps = 4/77 (5%)
Query 23 SSLDMNEWKKVYSNTQ-DLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLTSNQL 80
S + +++WK++YSNT+ + R VA+K WD D E S + +Y K E V+++T N++
Sbjct 264 SPMVLDDWKRLYSNTKSNFREVAIKGFWDMYDPEGYSLWFCDY-KYNDENMVSFVTLNKV 322
Query 81 GGFLQRVDNSMRKDSFA 97
GGFLQR+D + RK SF
Sbjct 323 GGFLQRMDLA-RKYSFG 338
> dre:195822 eef1g, db03h10, fk76a09, mg:db03h10, wu:fk76a09;
eukaryotic translation elongation factor 1 gamma; K03233 elongation
factor 1-gamma
Length=442
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 51/83 (61%), Gaps = 4/83 (4%)
Query 18 AQVVESSLDMNEWKKVYSNTQDLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLT 76
A + +SS M+E+K+ YSN +D VA+ + WDH D E S + EY + E T+ +++
Sbjct 286 AHLPKSSFVMDEFKRKYSN-EDTMTVALPYFWDHFDREGFSIWYAEY-RFPEELTMAFMS 343
Query 77 SNQLGGFLQRVDNSMRKDSFAVI 99
N + G QR+D +RK++FA +
Sbjct 344 CNLITGMFQRLDK-LRKNAFASV 365
> sce:YPL048W CAM1, TEF3; CPBP; K03233 elongation factor 1-gamma
Length=415
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 22 ESSLDMNEWKKVYSNTQDLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLTSNQL 80
+S+ +++WK+ YSN +D R VA+ W W+H + E S + + Y K E T+T++++N +
Sbjct 263 KSTFVLDDWKRKYSN-EDTRPVALPWFWEHYNPEEYSLWKVTY-KYNDELTLTFMSNNLV 320
Query 81 GGFLQRVDNSMR 92
GGF R+ S +
Sbjct 321 GGFFNRLSASTK 332
> sce:YKL081W TEF4, EFC1; Tef4p; K03233 elongation factor 1-gamma
Length=412
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 45/72 (62%), Gaps = 3/72 (4%)
Query 22 ESSLDMNEWKKVYSNTQDLRGVAMKWLWDHVD-EHCSFFLMEYEKIEGECTVTYLTSNQL 80
+S+ +++WK+ YSN D R VA+ W W+H + E S + + Y K E T+T++++N +
Sbjct 260 KSTFVLDDWKRKYSN-DDTRPVALPWFWEHYNPEEYSIWKVGY-KYNDELTLTFMSNNLV 317
Query 81 GGFLQRVDNSMR 92
GGF R+ S +
Sbjct 318 GGFFNRLSASTK 329
> xla:397861 eef1g-b, ef1g, gig35; eukaryotic translation elongation
factor 1 gamma; K03233 elongation factor 1-gamma
Length=437
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 56/99 (56%), Gaps = 4/99 (4%)
Query 2 QQQQQQQQQQQQQAAAAQVVESSLDMNEWKKVYSNTQDLRGVAMKWLWDHVD-EHCSFFL 60
+ ++ + + + A + +SS M+E+K+ YSN +D VA+ + W+H + E S +
Sbjct 265 ESEKALAAEPKSKDPYAHLPKSSFIMDEFKRKYSN-EDTLTVALPYFWEHFEKEGWSIWY 323
Query 61 MEYEKIEGECTVTYLTSNQLGGFLQRVDNSMRKDSFAVI 99
EY K E T T+++ N + G QR+D +RK +FA +
Sbjct 324 AEY-KFPEELTQTFMSCNLITGMFQRLDK-LRKTAFASV 360
> xla:379456 eef1g-a, MGC64329, eef1g, ef1g, gig35; eukaryotic
translation elongation factor 1 gamma; K03233 elongation factor
1-gamma
Length=436
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 54/99 (54%), Gaps = 4/99 (4%)
Query 2 QQQQQQQQQQQQQAAAAQVVESSLDMNEWKKVYSNTQDLRGVAMKWLWDHVD-EHCSFFL 60
+ ++ + + + A + +SS M+E+K+ YSN +D VA+ + W+H D E S +
Sbjct 264 ESEKALAAEPKSKDPYAHLPKSSFIMDEFKRKYSN-EDTLTVALPYFWEHFDKEGWSIWY 322
Query 61 MEYEKIEGECTVTYLTSNQLGGFLQRVDNSMRKDSFAVI 99
EY K E T +++ + + G QR+D +RK FA +
Sbjct 323 AEY-KFPEELTQAFMSCSLITGMFQRLDK-LRKTGFASV 359
> mmu:67160 Eef1g, 2610301D06Rik, AA407312, EF1G, MGC103354, MGC144723,
MGC144724; eukaryotic translation elongation factor
1 gamma; K03233 elongation factor 1-gamma
Length=437
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 64/125 (51%), Gaps = 29/125 (23%)
Query 1 QQQQQQQQQQQQQQAAAAQVVESSLD-------------------------MNEWKKVYS 35
+++Q+ Q ++++++ AAA E +D ++E+K+ YS
Sbjct 239 EEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAKDPFAHLPKSTFVLDEFKRKYS 298
Query 36 NTQDLRGVAMKWLWDHVDEH-CSFFLMEYEKIEGECTVTYLTSNQLGGFLQRVDNSMRKD 94
N +D VA+ + W+H D+ S + EY E E T T+++ N + G QR+D +RK+
Sbjct 299 N-EDTLSVALPYFWEHFDKDGWSLWYAEYRFPE-ELTQTFMSCNLITGMFQRLDK-LRKN 355
Query 95 SFAVI 99
+FA +
Sbjct 356 AFASV 360
> hsa:1937 EEF1G, EF1G, GIG35; eukaryotic translation elongation
factor 1 gamma; K03233 elongation factor 1-gamma
Length=437
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 64/125 (51%), Gaps = 29/125 (23%)
Query 1 QQQQQQQQQQQQQQAAAAQVVESSLD-------------------------MNEWKKVYS 35
+++Q+ Q ++++++ AAA E +D ++E+K+ YS
Sbjct 239 EEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAKDPFAHLPKSTFVLDEFKRKYS 298
Query 36 NTQDLRGVAMKWLWDHVDEH-CSFFLMEYEKIEGECTVTYLTSNQLGGFLQRVDNSMRKD 94
N +D VA+ + W+H D+ S + EY E E T T+++ N + G QR+D +RK+
Sbjct 299 N-EDTLSVALPYFWEHFDKDGWSLWYSEYRFPE-ELTQTFMSCNLITGMFQRLDK-LRKN 355
Query 95 SFAVI 99
+FA +
Sbjct 356 AFASV 360
Lambda K H
0.316 0.126 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40