bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
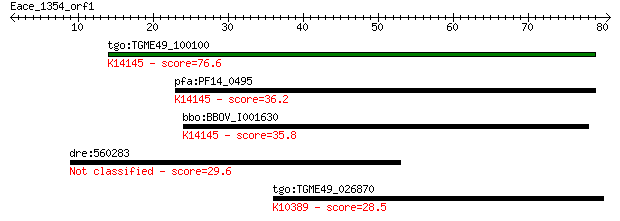
Query= Eace_1354_orf1
Length=80
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145... 76.6 2e-14
pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry n... 36.2 0.030
bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhopt... 35.8 0.033
dre:560283 cax1, MGC136271, si:dkey-180p18.2, wu:fc25f10, zgc:... 29.6 2.7
tgo:TGME49_026870 tubulin gamma chain, putative ; K10389 tubul... 28.5 5.2
> tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145
rhoptry neck protein 2
Length=1404
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 31/65 (47%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 14 LKTLEPFLMASNPLTTGHILTLLIGYIDRDAFFGSSPRKSFYNFTTLVGATGGESGILML 73
L +E F + NP T GH+LTL+I Y+D ++FFG+SP K F+++ +L + G +G ML
Sbjct 392 LAAIEIFRLGPNPYTIGHVLTLMIAYLDYESFFGASPSKPFHSWVSLAASAGNNTGFAML 451
Query 74 DEMCD 78
DEMCD
Sbjct 452 DEMCD 456
> pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry
neck protein 2
Length=2189
Score = 36.2 bits (82), Expect = 0.030, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 23 ASNPLTTGHILTLLIGYIDRDAFFGSSPRKSFYNFTTLVGATGGESGILMLDEMCD 78
ASN H L L + Y+ + +F ++ KSFY+ T++ A S ML+EMC+
Sbjct 1114 ASNIYLLAHFLVLSLAYLSYNEYF-TTGTKSFYSLPTILTANSDNS-FFMLNEMCN 1167
> bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhoptry
neck protein 2
Length=1365
Score = 35.8 bits (81), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 27/55 (49%), Gaps = 6/55 (10%)
Query 24 SNPLTTGHILTLLIGYIDRDAFFGSSPRKSFYNFTTLVGATGGESGIL-MLDEMC 77
+NP GH+ T ++ Y + FF + FY + LV SG L MLD MC
Sbjct 366 NNPFLLGHLATNMLAYTQYNMFFAGGSGRPFYTWLDLVS-----SGNLDMLDRMC 415
> dre:560283 cax1, MGC136271, si:dkey-180p18.2, wu:fc25f10, zgc:136271;
cation/H+ exchanger protein 1
Length=764
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 3/44 (6%)
Query 9 YSKRKLKTLEPFLMASNPLTTGHILTLLIGYIDRDAFFGSSPRK 52
Y K + + F + PL I+TL+IGY+DR+ +F SS K
Sbjct 350 YYKYTVDGINVFAVNLLPLV---IITLIIGYMDRENYFVSSEVK 390
> tgo:TGME49_026870 tubulin gamma chain, putative ; K10389 tubulin
gamma
Length=455
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 36 LIGYIDRDAFFGSSPRKSFYNFTTLVGATGGESGILMLDEMCDR 79
L+ IDR+A GS + F ++ G TG G +L+ +CDR
Sbjct 120 LLEMIDREAD-GSESLEGFVLCHSIAGGTGSGMGSYLLEALCDR 162
Lambda K H
0.321 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2058492688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40