bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1395_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
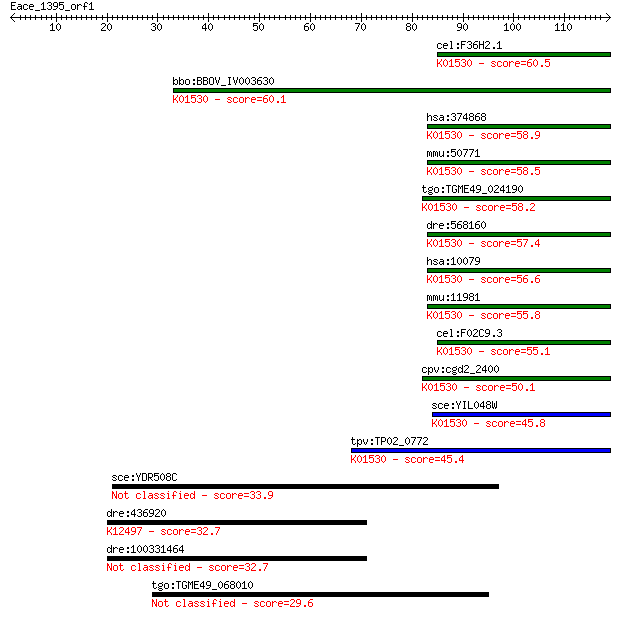
cel:F36H2.1 tat-5; Transbilayer Amphipath Transporters (subfam... 60.5 1e-09
bbo:BBOV_IV003630 21.m02979; phospholipid-translocating P-type... 60.1 2e-09
hsa:374868 ATP9B, ATPASEP, ATPIIB, DKFZp686H2093, FLJ46612, MG... 58.9 4e-09
mmu:50771 Atp9b, AA934181, Atpc2b, IIb, MMR; ATPase, class II,... 58.5 5e-09
tgo:TGME49_024190 phospholipid-transporting ATPase, P-type, pu... 58.2 7e-09
dre:568160 atp9b, si:ch211-198c22.1; ATPase, class II, type 9B... 57.4 1e-08
hsa:10079 ATP9A, ATPIIA, KIAA0611; ATPase, class II, type 9A (... 56.6 2e-08
mmu:11981 Atp9a, IIa, mKIAA0611; ATPase, class II, type 9A (EC... 55.8 3e-08
cel:F02C9.3 tat-6; Transbilayer Amphipath Transporters (subfam... 55.1 5e-08
cpv:cgd2_2400 ATPase, class II, type 9B ; K01530 phospholipid-... 50.1 2e-06
sce:YIL048W NEO1; Neo1p (EC:3.6.3.1); K01530 phospholipid-tran... 45.8 3e-05
tpv:TP02_0772 phospholipid-transporting ATPase; K01530 phospho... 45.4 4e-05
sce:YDR508C GNP1; Gnp1p 33.9 0.13
dre:436920 zgc:92020; K12497 early growth response protein 3 32.7
dre:100331464 early growth response 3-like 32.7 0.29
tgo:TGME49_068010 hypothetical protein 29.6 2.7
> cel:F36H2.1 tat-5; Transbilayer Amphipath Transporters (subfamily
IV P-type ATPase) family member (tat-5); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1035
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 30/34 (88%), Gaps = 0/34 (0%)
Query 85 LGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
L +E VLWANTVVA+GT +GIV+YTGRETRS MN
Sbjct 264 LNVENVLWANTVVASGTAVGIVVYTGRETRSVMN 297
> bbo:BBOV_IV003630 21.m02979; phospholipid-translocating P-type
ATPase (EC:3.6.3.1); K01530 phospholipid-translocating ATPase
[EC:3.6.3.1]
Length=1127
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 35/89 (39%), Positives = 49/89 (55%), Gaps = 6/89 (6%)
Query 33 DDQPTDKTSSSGAATPAAGTAATAAA---AAAAAGPGDTMVDVSGGGCSYCAVEPLGLEQ 89
D D ++ T A T+ + A + A G D V+ + GG VEPL ++
Sbjct 268 DSMRLDTSTLKDFGTQAEMTSVSYVALETKSKAEGDADCDVEEAPGGV---IVEPLTVDN 324
Query 90 VLWANTVVAAGTVIGIVIYTGRETRSSMN 118
LW NT+VAAGTV G+VIY G+E R+S+N
Sbjct 325 ALWMNTIVAAGTVYGLVIYIGKEVRASLN 353
> hsa:374868 ATP9B, ATPASEP, ATPIIB, DKFZp686H2093, FLJ46612,
MGC150650, MGC150651, MGC61572, NEO1L; ATPase, class II, type
9B (EC:3.6.3.1); K01530 phospholipid-translocating ATPase
[EC:3.6.3.1]
Length=1147
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 25/36 (69%), Positives = 31/36 (86%), Gaps = 0/36 (0%)
Query 83 EPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
E L +E LWA+T+VA+GTVIG+VIYTG+ETRS MN
Sbjct 327 ESLSIENTLWASTIVASGTVIGVVIYTGKETRSVMN 362
> mmu:50771 Atp9b, AA934181, Atpc2b, IIb, MMR; ATPase, class II,
type 9B (EC:3.6.3.1); K01530 phospholipid-translocating ATPase
[EC:3.6.3.1]
Length=1146
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 25/36 (69%), Positives = 31/36 (86%), Gaps = 0/36 (0%)
Query 83 EPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
E L +E LWA+T+VA+GTVIG+VIYTG+ETRS MN
Sbjct 326 ESLSIENTLWASTIVASGTVIGVVIYTGKETRSVMN 361
> tgo:TGME49_024190 phospholipid-transporting ATPase, P-type,
putative (EC:3.6.3.1); K01530 phospholipid-translocating ATPase
[EC:3.6.3.1]
Length=1138
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 25/37 (67%), Positives = 33/37 (89%), Gaps = 0/37 (0%)
Query 82 VEPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
VE L L+ VLWANTVVA+G+V+G+ IYTG+ETR++MN
Sbjct 331 VEGLTLDNVLWANTVVASGSVLGLAIYTGKETRATMN 367
> dre:568160 atp9b, si:ch211-198c22.1; ATPase, class II, type
9B (EC:3.6.3.1); K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1108
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/36 (69%), Positives = 30/36 (83%), Gaps = 0/36 (0%)
Query 83 EPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
E L +E LWA+TVVA+GTVIG+VIYTG+E RS MN
Sbjct 297 ESLSIENTLWASTVVASGTVIGVVIYTGKEMRSVMN 332
> hsa:10079 ATP9A, ATPIIA, KIAA0611; ATPase, class II, type 9A
(EC:3.6.3.1); K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1047
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 24/36 (66%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 83 EPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
E L +E LWA TVVA+GTV+G+V+YTGRE RS MN
Sbjct 250 ESLSIENTLWAGTVVASGTVVGVVLYTGRELRSVMN 285
> mmu:11981 Atp9a, IIa, mKIAA0611; ATPase, class II, type 9A (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1047
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/36 (63%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 83 EPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
E L +E LWA TV+A+GTV+G+V+YTGRE RS MN
Sbjct 250 ESLSIENTLWAGTVIASGTVVGVVLYTGRELRSVMN 285
> cel:F02C9.3 tat-6; Transbilayer Amphipath Transporters (subfamily
IV P-type ATPase) family member (tat-6); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1064
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 24/34 (70%), Positives = 29/34 (85%), Gaps = 0/34 (0%)
Query 85 LGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
L +E VLWANTVV++GT IG+V+YTG ETRS MN
Sbjct 293 LDVENVLWANTVVSSGTAIGVVVYTGCETRSVMN 326
> cpv:cgd2_2400 ATPase, class II, type 9B ; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1291
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 21/37 (56%), Positives = 28/37 (75%), Gaps = 0/37 (0%)
Query 82 VEPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
VEPL L+ +WAN+V+ G + G+VIYTG E+RS MN
Sbjct 288 VEPLMLDNTIWANSVLTCGKIYGLVIYTGIESRSLMN 324
> sce:YIL048W NEO1; Neo1p (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1151
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 20/36 (55%), Positives = 27/36 (75%), Gaps = 1/36 (2%)
Query 84 PLGLEQVLWANTVVAA-GTVIGIVIYTGRETRSSMN 118
PL ++ LWANTV+A+ G I V+YTGR+TR +MN
Sbjct 361 PLSVDNTLWANTVLASSGFCIACVVYTGRDTRQAMN 396
> tpv:TP02_0772 phospholipid-transporting ATPase; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1280
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 68 TMVDVSGGGCSYCAVEPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
T +DV S VE L ++ ++W N++VA+GT+ G+ IY G++ R+ +N
Sbjct 335 TSIDVESKLESKRIVESLNVDNIVWMNSIVASGTIHGLTIYIGKDARACLN 385
> sce:YDR508C GNP1; Gnp1p
Length=663
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 18/77 (23%), Positives = 35/77 (45%), Gaps = 1/77 (1%)
Query 21 QQQQQKQQQQQRDDQPTDKTSSSGAATPAAGTAATAAAAAAAAGPGDTMVDVS-GGGCSY 79
Q+Q+QKQ+ ++ +P S G + AGPG ++ + G C Y
Sbjct 136 QEQEQKQENLKKSIKPRHTVMMSLGTGIGTGLLVGNSKVLNNAGPGGLIIGYAIMGSCVY 195
Query 80 CAVEPLGLEQVLWANTV 96
C ++ G V++++ +
Sbjct 196 CIIQACGELAVIYSDLI 212
> dre:436920 zgc:92020; K12497 early growth response protein 3
Length=422
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 4/51 (7%)
Query 20 QQQQQQKQQQQQRDDQPTDKTSSSGAATPAAGTAATAAAAAAAAGPGDTMV 70
++++ K +Q+D +PTDK SS+G A G + ++ +AGP T +
Sbjct 369 ERKRHAKVHLKQKDKKPTDKGSSAG----AGGNHTSPPSSCGSAGPSSTNI 415
> dre:100331464 early growth response 3-like
Length=422
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 4/51 (7%)
Query 20 QQQQQQKQQQQQRDDQPTDKTSSSGAATPAAGTAATAAAAAAAAGPGDTMV 70
++++ K +Q+D +PTDK SS+G A G + ++ +AGP T +
Sbjct 369 ERKRHAKVHLKQKDKKPTDKGSSAG----AGGNHTSPPSSCGSAGPSSTNI 415
> tgo:TGME49_068010 hypothetical protein
Length=4032
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 2/68 (2%)
Query 29 QQQRDDQPTDKT--SSSGAATPAAGTAATAAAAAAAAGPGDTMVDVSGGGCSYCAVEPLG 86
++ RDD+P +K S+ P+A + A++ A AAA P D G C+ P+
Sbjct 3495 EEPRDDRPEEKRLASADREPGPSANSPASSKDAPAAALPPGEKADELGDSCAASTFPPIP 3554
Query 87 LEQVLWAN 94
WA
Sbjct 3555 SFVAAWAE 3562
Lambda K H
0.309 0.121 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40