bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1397_orf1
Length=138
Score E
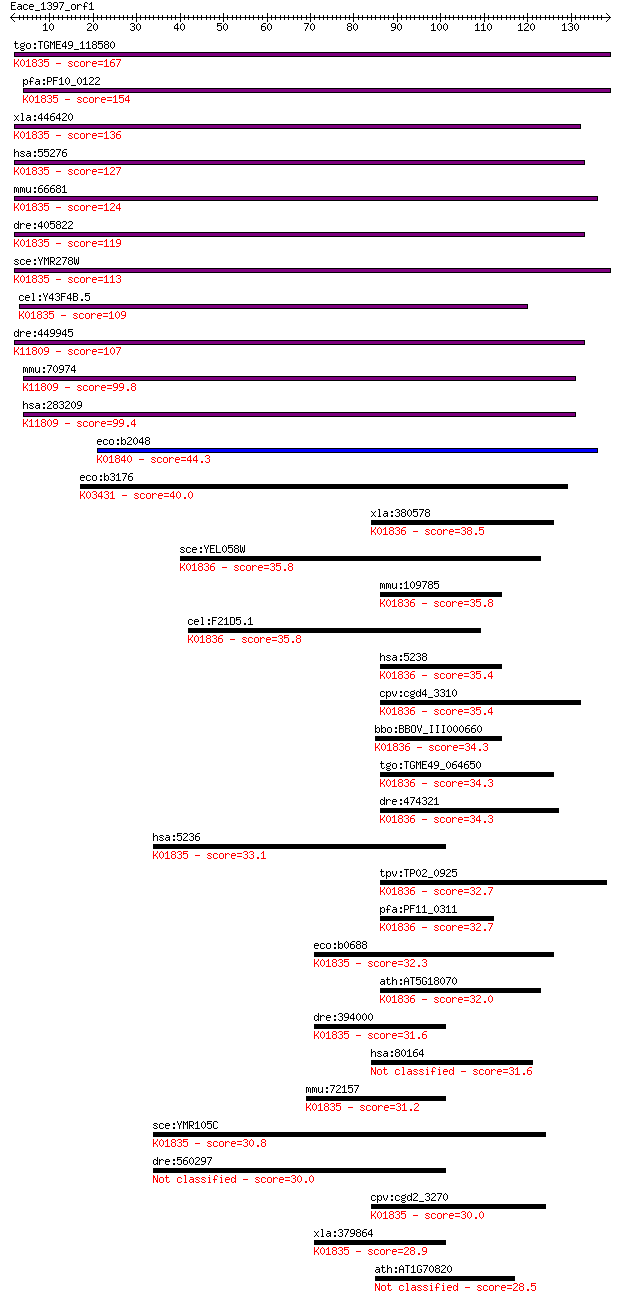
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118580 phosphoglucomutase, putative (EC:5.4.2.8); K... 167 7e-42
pfa:PF10_0122 phosphoglucomutase, putative; K01835 phosphogluc... 154 6e-38
xla:446420 pgm2l1, MGC83889; phosphoglucomutase 2-like 1 (EC:2... 136 2e-32
hsa:55276 PGM2, FLJ10983; phosphoglucomutase 2 (EC:5.4.2.2 5.4... 127 1e-29
mmu:66681 Pgm1, 3230402E02Rik, Pgm-1, Pgm2; phosphoglucomutase... 124 1e-28
dre:405822 pgm2, MGC77113, zgc:77113; phosphoglucomutase 2 (EC... 119 3e-27
sce:YMR278W PGM3; Pgm3p; K01835 phosphoglucomutase [EC:5.4.2.2] 113 2e-25
cel:Y43F4B.5 hypothetical protein; K01835 phosphoglucomutase [... 109 2e-24
dre:449945 pgm2l1, im:7140576, zgc:198285; phosphoglucomutase ... 107 1e-23
mmu:70974 Pgm2l1, 4931406N15Rik, AI553438, BM32A; phosphogluco... 99.8 2e-21
hsa:283209 PGM2L1, BM32A, FLJ32029; phosphoglucomutase 2-like ... 99.4 3e-21
eco:b2048 cpsG, ECK2042, JW2033, manB, rfbK; phosphomannomutas... 44.3 1e-04
eco:b3176 glmM, ECK3165, JW3143, mrsA, yhbF; phosphoglucosamin... 40.0 0.002
xla:380578 pgm3, MGC69105; phosphoglucomutase 3 (EC:5.4.2.2); ... 38.5 0.006
sce:YEL058W PCM1, AGM1; Essential N-acetylglucosamine-phosphat... 35.8 0.036
mmu:109785 Pgm3, 2810473H05Rik, Agm1, BB187688, C77933, PAGM, ... 35.8 0.041
cel:F21D5.1 hypothetical protein; K01836 phosphoacetylglucosam... 35.8 0.044
hsa:5238 PGM3, AGM1, DKFZp434B187, FLJ11614, FLJ13623, PAGM, P... 35.4 0.050
cpv:cgd4_3310 phosphoacetyl glucosamine mutase ; K01836 phosph... 35.4 0.052
bbo:BBOV_III000660 phosphoglucomutase; K01836 phosphoacetylglu... 34.3 0.10
tgo:TGME49_064650 hypothetical protein ; K01836 phosphoacetylg... 34.3 0.13
dre:474321 pgm3, wu:fc08c11, zgc:91932; phosphoglucomutase 3 (... 34.3 0.13
hsa:5236 PGM1, GSD14; phosphoglucomutase 1 (EC:5.4.2.2); K0183... 33.1 0.29
tpv:TP02_0925 N-acetylglucosamine-phosphate mutase; K01836 pho... 32.7 0.33
pfa:PF11_0311 N-acetyl glucosamine phosphate mutase, putative;... 32.7 0.35
eco:b0688 pgm, blu, ECK0676, JW0675, pgmA; phosphoglucomutase ... 32.3 0.51
ath:AT5G18070 DRT101; DRT101 (DNA-DAMAGE-REPAIR/TOLERATION 101... 32.0 0.60
dre:394000 pgm1, MGC63718, zgc:63718; phosphoglucomutase 1 (EC... 31.6 0.72
hsa:80164 hypothetical protein FLJ22184 31.6 0.86
mmu:72157 Pgm2, 2610020G18Rik, AA407108, AI098105, Pgm-2, Pgm1... 31.2 0.93
sce:YMR105C PGM2, GAL5; Pgm2p (EC:5.4.2.2); K01835 phosphogluc... 30.8 1.2
dre:560297 novel protein similar to vertebrate phosphoglucomut... 30.0 2.0
cpv:cgd2_3270 phosphoglucomutase, tandemly duplicated gene (EC... 30.0 2.1
xla:379864 pgm1, MGC53760, pgm2; phosphoglucomutase 1 (EC:5.4.... 28.9 5.1
ath:AT1G70820 phosphoglucomutase, putative / glucose phosphomu... 28.5 6.4
> tgo:TGME49_118580 phosphoglucomutase, putative (EC:5.4.2.8);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=597
Score = 167 bits (424), Expect = 7e-42, Method: Composition-based stats.
Identities = 78/137 (56%), Positives = 98/137 (71%), Gaps = 1/137 (0%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRHNSERFSHLAAATFLSRGFR 61
SR+NDVTIQQ +QG +L+ G D RG++IG D RHNS RF+ L AA FLS+GFR
Sbjct 69 SRMNDVTIQQTTQGYCAFLVDVFGED-GKDRGVVIGFDARHNSRRFAQLTAAVFLSKGFR 127
Query 62 VYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADIAN 121
V L VHTP+V + V+ NC+AGI+ITASHNPK NGYKVY NGAQIIPP+D++I+
Sbjct 128 VQLFSDIVHTPMVPYTVVAANCIAGIMITASHNPKADNGYKVYAANGAQIIPPMDSEISA 187
Query 122 AIQNNQSLWPEVEEVFD 138
I +N W +V+E FD
Sbjct 188 FINSNLDFWSDVDEYFD 204
> pfa:PF10_0122 phosphoglucomutase, putative; K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=593
Score = 154 bits (390), Expect = 6e-38, Method: Composition-based stats.
Identities = 71/135 (52%), Positives = 93/135 (68%), Gaps = 0/135 (0%)
Query 4 LNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRHNSERFSHLAAATFLSRGFRVY 63
+N VTI Q +QGL YLI +G ++C RGII G D R++SE F+H+AA+ LS+GFRVY
Sbjct 67 MNVVTIMQTTQGLCSYLINTYGLNLCKNRGIIFGFDGRYHSESFAHVAASVCLSKGFRVY 126
Query 64 LCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADIANAI 123
L V TP++ + L NC+ G+++TASHNPK NGYKVY NGAQIIPPVD +I+N I
Sbjct 127 LFAQTVATPILCYSNLKKNCLCGVMVTASHNPKLDNGYKVYASNGAQIIPPVDKNISNCI 186
Query 124 QNNQSLWPEVEEVFD 138
NN W + E +
Sbjct 187 LNNLEPWKDAYEYLN 201
> xla:446420 pgm2l1, MGC83889; phosphoglucomutase 2-like 1 (EC:2.7.1.106);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=610
Score = 136 bits (342), Expect = 2e-32, Method: Composition-based stats.
Identities = 68/136 (50%), Positives = 93/136 (68%), Gaps = 8/136 (5%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRH------NSERFSHLAAATF 55
S++ND+TI Q +QG YL +++ +D+ RG++IG D R +S+RF+ LAA TF
Sbjct 69 SQMNDLTIIQTTQGFCRYL-EKNISDL-KERGVVIGYDARAHPASGGSSKRFARLAATTF 126
Query 56 LSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPV 115
+S+G +VY+ TP V + V HLN AGI++TASHNPK+ NGYKVY ENGAQIIPP
Sbjct 127 VSQGIKVYMFSDITPTPFVPYAVTHLNLCAGIMVTASHNPKQDNGYKVYWENGAQIIPPH 186
Query 116 DADIANAIQNNQSLWP 131
D+ IA +I+ N WP
Sbjct 187 DSGIAQSIEKNLEPWP 202
> hsa:55276 PGM2, FLJ10983; phosphoglucomutase 2 (EC:5.4.2.2 5.4.2.7);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=612
Score = 127 bits (319), Expect = 1e-29, Method: Composition-based stats.
Identities = 69/137 (50%), Positives = 87/137 (63%), Gaps = 8/137 (5%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRH------NSERFSHLAAATF 55
SR+ND+TI Q +QG YL K+ +D+ +GI+I D R +S RF+ LAA TF
Sbjct 71 SRMNDLTIIQTTQGFCRYLEKQF-SDL-KQKGIVISFDARAHPSSGGSSRRFARLAATTF 128
Query 56 LSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPV 115
+S+G VYL TP V F V HL AGI+ITASHNPK+ NGYKVY +NGAQII P
Sbjct 129 ISQGIPVYLFSDITPTPFVPFTVSHLKLCAGIMITASHNPKQDNGYKVYWDNGAQIISPH 188
Query 116 DADIANAIQNNQSLWPE 132
D I+ AI+ N WP+
Sbjct 189 DKGISQAIEENLEPWPQ 205
> mmu:66681 Pgm1, 3230402E02Rik, Pgm-1, Pgm2; phosphoglucomutase
1 (EC:5.4.2.2 5.4.2.7); K01835 phosphoglucomutase [EC:5.4.2.2]
Length=620
Score = 124 bits (310), Expect = 1e-28, Method: Composition-based stats.
Identities = 67/140 (47%), Positives = 87/140 (62%), Gaps = 8/140 (5%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRH------NSERFSHLAAATF 55
SR+ND+TI Q +QG YL K+ +D+ RG++I D R +S RF+ LAA F
Sbjct 79 SRMNDLTIIQTTQGFCRYLEKQF-SDL-KQRGVVISFDARAHPASGGSSRRFARLAATAF 136
Query 56 LSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPV 115
+++G VYL TP V + V HL AGI+ITASHNPK+ NGYKVY +NGAQII P
Sbjct 137 ITQGVPVYLFSDITPTPFVPYTVSHLKLCAGIMITASHNPKQDNGYKVYWDNGAQIISPH 196
Query 116 DADIANAIQNNQSLWPEVEE 135
D I+ AI+ N WP+ E
Sbjct 197 DRGISQAIEENLEPWPQAWE 216
> dre:405822 pgm2, MGC77113, zgc:77113; phosphoglucomutase 2 (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=611
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 68/137 (49%), Positives = 84/137 (61%), Gaps = 8/137 (5%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCR------HNSERFSHLAAATF 55
S +ND+TI Q +QG YL + D+ RG++IG D R +S+RF+ LAA
Sbjct 69 SCMNDLTIIQTTQGFCAYL-EECFVDL-KQRGVVIGFDARAHPPSGGSSKRFACLAACVL 126
Query 56 LSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPV 115
+SRG V+L TP V F V HL AGI++TASHNPK+ NGYKVY NGAQIIPP
Sbjct 127 MSRGVPVHLFSDITPTPYVPFAVSHLGLCAGIMVTASHNPKQDNGYKVYWANGAQIIPPH 186
Query 116 DADIANAIQNNQSLWPE 132
D IA AI+ N WPE
Sbjct 187 DKGIAAAIEQNLEPWPE 203
> sce:YMR278W PGM3; Pgm3p; K01835 phosphoglucomutase [EC:5.4.2.2]
Length=622
Score = 113 bits (282), Expect = 2e-25, Method: Composition-based stats.
Identities = 61/140 (43%), Positives = 84/140 (60%), Gaps = 7/140 (5%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRHNSERFSHLAAATFLSRGFR 61
SR+N + + QASQGLA Y+ ++ ++ + ++G D R +S+ F+ AA FL +GF+
Sbjct 69 SRMNTLVVIQASQGLATYVRQQFPDNLVA----VVGHDHRFHSKEFARATAAAFLLKGFK 124
Query 62 VYLC---RSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDAD 118
V+ VHTPLV F V L G++ITASHNPK NGYKVY NG QIIPP D
Sbjct 125 VHYLNPDHEFVHTPLVPFAVDKLKASVGVMITASHNPKMDNGYKVYYSNGCQIIPPHDHA 184
Query 119 IANAIQNNQSLWPEVEEVFD 138
I+++I N W V + D
Sbjct 185 ISDSIDANLEPWANVWDFDD 204
> cel:Y43F4B.5 hypothetical protein; K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=595
Score = 109 bits (273), Expect = 2e-24, Method: Composition-based stats.
Identities = 55/117 (47%), Positives = 73/117 (62%), Gaps = 3/117 (2%)
Query 3 RLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRHNSERFSHLAAATFLSRGFRV 62
RLND+TI Q + G A +++ +G G+ IG D R+NS RF+ L+A F+ V
Sbjct 65 RLNDLTIIQITHGFARHMLNVYGQ---PKNGVAIGFDGRYNSRRFAELSANVFVRNNIPV 121
Query 63 YLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADI 119
YL TP+V++ + L C AG++ITASHNPKE NGYK Y NGAQII P D +I
Sbjct 122 YLFSEVSPTPVVSWATIKLGCDAGLIITASHNPKEDNGYKAYWSNGAQIIGPHDTEI 178
> dre:449945 pgm2l1, im:7140576, zgc:198285; phosphoglucomutase
2-like 1 (EC:2.7.1.106); K11809 glucose-1,6-bisphosphate synthase
[EC:2.7.1.106]
Length=619
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 61/137 (44%), Positives = 81/137 (59%), Gaps = 8/137 (5%)
Query 2 SRLNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRH------NSERFSHLAAATF 55
+R+ND+TI Q++QGL YL K D+ TRG+++G D R SER + L AA
Sbjct 78 ARINDLTIIQSTQGLYKYLAKCF-PDL-KTRGLVVGYDTRAQASSGCTSERLAKLTAAVM 135
Query 56 LSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPV 115
L + VYL + V TP V + V+ AG++ITASHN KE NGYKVY NGAQI P
Sbjct 136 LCKDVPVYLFSTYVPTPFVPYAVMKYGAAAGVMITASHNRKEDNGYKVYWHNGAQIASPH 195
Query 116 DADIANAIQNNQSLWPE 132
D +I + I+ + W E
Sbjct 196 DKEILHCIEESAEPWAE 212
> mmu:70974 Pgm2l1, 4931406N15Rik, AI553438, BM32A; phosphoglucomutase
2-like 1 (EC:2.7.1.106); K11809 glucose-1,6-bisphosphate
synthase [EC:2.7.1.106]
Length=621
Score = 99.8 bits (247), Expect = 2e-21, Method: Composition-based stats.
Identities = 57/133 (42%), Positives = 77/133 (57%), Gaps = 8/133 (6%)
Query 4 LNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRH------NSERFSHLAAATFLS 57
+ND+T+ Q++QG+ YL +R +D RG ++G D R +S+R + L AA L+
Sbjct 83 INDLTVIQSTQGMYKYL-ERCFSDF-KQRGFVVGYDTRGQVTSSCSSQRLAKLTAAVLLA 140
Query 58 RGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDA 117
+ VYL V TP V + V L VAG++ITASHN KE NGYKVY E GAQI P D
Sbjct 141 KDIPVYLFSRYVPTPFVPYAVQELKAVAGVMITASHNRKEDNGYKVYWETGAQITSPHDK 200
Query 118 DIANAIQNNQSLW 130
+I I+ W
Sbjct 201 EILKCIEECVEPW 213
> hsa:283209 PGM2L1, BM32A, FLJ32029; phosphoglucomutase 2-like
1 (EC:2.7.1.106); K11809 glucose-1,6-bisphosphate synthase
[EC:2.7.1.106]
Length=622
Score = 99.4 bits (246), Expect = 3e-21, Method: Composition-based stats.
Identities = 57/133 (42%), Positives = 77/133 (57%), Gaps = 8/133 (6%)
Query 4 LNDVTIQQASQGLADYLIKRHGADICSTRGIIIGRDCRH------NSERFSHLAAATFLS 57
+ND+T+ Q++QG+ YL +R +D RG ++G D R +S+R + L AA L+
Sbjct 83 INDLTVIQSTQGMYKYL-ERCFSDF-KQRGFVVGYDTRGQVTSSCSSQRLAKLTAAVLLA 140
Query 58 RGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDA 117
+ VYL V TP V + V L VAG++ITASHN KE NGYKVY E GAQI P D
Sbjct 141 KDVPVYLFSRYVPTPFVPYAVQKLKAVAGVMITASHNRKEDNGYKVYWETGAQITSPHDK 200
Query 118 DIANAIQNNQSLW 130
+I I+ W
Sbjct 201 EILKCIEECVEPW 213
> eco:b2048 cpsG, ECK2042, JW2033, manB, rfbK; phosphomannomutase
(EC:5.4.2.8); K01840 phosphomannomutase [EC:5.4.2.8]
Length=456
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 38/120 (31%), Positives = 53/120 (44%), Gaps = 12/120 (10%)
Query 21 IKRHGADICSTRGIIIGRDCRHNSERFSHLAAATFLSRGFRVYLCRSAVHTPLVAFGVLH 80
I R + + I++G D R SE A G V L T + F H
Sbjct 28 IGRAYGEFLKPKTIVLGGDVRLTSETLKLALAKGLQDAGVDV-LDIGMSGTEEIYFATFH 86
Query 81 LNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADIANAIQNNQSL-----WPEVEE 135
L GI +TASHNP +YNG K+ E GA+ P+ D +++ Q L +P V+E
Sbjct 87 LGVDGGIEVTASHNPMDYNGMKLVRE-GAR---PISGD--TGLRDVQRLAEANDFPPVDE 140
> eco:b3176 glmM, ECK3165, JW3143, mrsA, yhbF; phosphoglucosamine
mutase (EC:5.4.2.10); K03431 phosphoglucosamine mutase [EC:5.4.2.10]
Length=445
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 34/112 (30%), Positives = 50/112 (44%), Gaps = 6/112 (5%)
Query 17 ADYLIKRHGADICSTRGIIIGRDCRHNSERFSHLAAATFLSRGFRVYLCRSAVHTPLVAF 76
A ++ RHG +R IIIG+D R + A + G + TP VA+
Sbjct 33 AGKVLARHG-----SRKIIIGKDTRISGYMLESALEAGLAAAGLSALFT-GPMPTPAVAY 86
Query 77 GVLHLNCVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADIANAIQNNQS 128
AGIVI+ASHNP NG K + +G ++ V+ I ++ S
Sbjct 87 LTRTFRAEAGIVISASHNPFYDNGIKFFSIDGTKLPDAVEEAIEAEMEKEIS 138
> xla:380578 pgm3, MGC69105; phosphoglucomutase 3 (EC:5.4.2.2);
K01836 phosphoacetylglucosamine mutase [EC:5.4.2.3]
Length=542
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 29/45 (64%), Gaps = 3/45 (6%)
Query 84 VAGIVITASHNPKEYNGYKVYDENGAQIIPPVD---ADIANAIQN 125
V G+++TASHNP+E NG K+ D G ++ + ++ANA QN
Sbjct 56 VIGVMVTASHNPEEDNGVKLVDPMGEMLVQEWEVYATNLANAEQN 100
> sce:YEL058W PCM1, AGM1; Essential N-acetylglucosamine-phosphate
mutase; converts GlcNAc-6-P to GlcNAc-1-P, which is a precursor
for the biosynthesis of chitin and for the formation
of N-glycosylated mannoproteins and glycosylphosphatidylinositol
anchors (EC:5.4.2.3); K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=557
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 18/93 (19%)
Query 40 CRHNSERFSHLAAATFLSRGFRVYLCRSAVHTPLVAFGVLHL-------NCVAGIVITAS 92
CR + +FS+ A GFR L ++ + T + + G+L + G++ITAS
Sbjct 16 CRTKNVQFSYGTA------GFRT-LAKN-LDTVMFSTGILAVLRSLKLQGQYVGVMITAS 67
Query 93 HNPKEYNGYKVYDENGAQIIP---PVDADIANA 122
HNP + NG K+ + +G+ ++ P +ANA
Sbjct 68 HNPYQDNGVKIVEPDGSMLLATWEPYAMQLANA 100
> mmu:109785 Pgm3, 2810473H05Rik, Agm1, BB187688, C77933, PAGM,
Pgm-3; phosphoglucomutase 3 (EC:5.4.2.3); K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=501
Score = 35.8 bits (81), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQIIP 113
G+++TASHNP+E NG K+ D G + P
Sbjct 58 GVMVTASHNPEEDNGVKLVDPLGEMLAP 85
> cel:F21D5.1 hypothetical protein; K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=550
Score = 35.8 bits (81), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 33/68 (48%), Gaps = 4/68 (5%)
Query 42 HNSERFSH-LAAATFLSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVITASHNPKEYNG 100
+ E+FS+ A F S + R A L A LN G++ITASHNP NG
Sbjct 25 QDDEKFSYGTAGFRFKSEKLPFIVFRCAYVASLRA---RQLNSAIGVMITASHNPSCDNG 81
Query 101 YKVYDENG 108
K+ D +G
Sbjct 82 VKLVDPSG 89
> hsa:5238 PGM3, AGM1, DKFZp434B187, FLJ11614, FLJ13623, PAGM,
PGM_3; phosphoglucomutase 3 (EC:5.4.2.3); K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=570
Score = 35.4 bits (80), Expect = 0.050, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQIIP 113
G+++TASHNP+E NG K+ D G + P
Sbjct 86 GVMVTASHNPEEDNGVKLVDPLGEMLAP 113
> cpv:cgd4_3310 phosphoacetyl glucosamine mutase ; K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=643
Score = 35.4 bits (80), Expect = 0.052, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQIIPPVDADIANAIQNNQSLWP 131
G+VITASHNP + NG K+ D G+ + + NA+ + + P
Sbjct 66 GVVITASHNPIQDNGVKIVDVTGSMLNKDFEQICFNAVNHEDASNP 111
> bbo:BBOV_III000660 phosphoglucomutase; K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=596
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 85 AGIVITASHNPKEYNGYKVYDENGAQIIP 113
AG+VITASHN E NG K+ + NG + P
Sbjct 99 AGVVITASHNGPEDNGIKILENNGNLLDP 127
> tgo:TGME49_064650 hypothetical protein ; K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=985
Score = 34.3 bits (77), Expect = 0.13, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQIIPPVDADIANAIQN 125
G +ITASHNP E NG K+ +G ++PPV A + N
Sbjct 112 GCMITASHNPVEDNGVKLVGRDGC-LLPPVWERYAETLVN 150
> dre:474321 pgm3, wu:fc08c11, zgc:91932; phosphoglucomutase 3
(EC:5.4.2.2); K01836 phosphoacetylglucosamine mutase [EC:5.4.2.3]
Length=545
Score = 34.3 bits (77), Expect = 0.13, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQIIPPVD---ADIANAIQNN 126
G+++TASHNP+E NG K+ D G + + +ANA Q++
Sbjct 59 GVMVTASHNPEEDNGVKLIDPMGEMVAATWEEYATQLANAEQDH 102
> hsa:5236 PGM1, GSD14; phosphoglucomutase 1 (EC:5.4.2.2); K01835
phosphoglucomutase [EC:5.4.2.2]
Length=580
Score = 33.1 bits (74), Expect = 0.29, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 36/71 (50%), Gaps = 6/71 (8%)
Query 34 IIIGRDCRHNS----ERFSHLAAATFLSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVI 89
+++G D R+ + E +AAA + R V + TP V+ + + + GI++
Sbjct 75 LVVGGDGRYFNKSAIETIVQMAAANGIGR--LVIGQNGILSTPAVSCIIRKIKAIGGIIL 132
Query 90 TASHNPKEYNG 100
TASHNP NG
Sbjct 133 TASHNPGGPNG 143
> tpv:TP02_0925 N-acetylglucosamine-phosphate mutase; K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=630
Score = 32.7 bits (73), Expect = 0.33, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQ---IIPPVDADIANAIQNNQSLWPEVEEVF 137
GIV+TASHNP NG K++ +G + P+ N + QS EV F
Sbjct 76 GIVVTASHNPCSDNGIKLFSPSGRTLECVWEPIFTSFVNTRNSIQSALYEVFTSF 130
> pfa:PF11_0311 N-acetyl glucosamine phosphate mutase, putative;
K01836 phosphoacetylglucosamine mutase [EC:5.4.2.3]
Length=940
Score = 32.7 bits (73), Expect = 0.35, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 86 GIVITASHNPKEYNGYKVYDENGAQI 111
GI+ITASHNP + NG K+ +G I
Sbjct 219 GIIITASHNPHDENGIKIIGVDGKYI 244
> eco:b0688 pgm, blu, ECK0676, JW0675, pgmA; phosphoglucomutase
(EC:5.4.2.2); K01835 phosphoglucomutase [EC:5.4.2.2]
Length=546
Score = 32.3 bits (72), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 28/60 (46%), Gaps = 9/60 (15%)
Query 71 TPLVAFGVLHLN-----CVAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADIANAIQN 125
TP V+ +L N GIVIT SHNP E G K NG P D ++ +++
Sbjct 120 TPAVSNAILVHNKKGGPLADGIVITPSHNPPEDGGIKYNPPNGG----PADTNVTKVVED 175
> ath:AT5G18070 DRT101; DRT101 (DNA-DAMAGE-REPAIR/TOLERATION 101);
intramolecular transferase, phosphotransferases / magnesium
ion binding (EC:5.4.2.3); K01836 phosphoacetylglucosamine
mutase [EC:5.4.2.3]
Length=556
Score = 32.0 bits (71), Expect = 0.60, Method: Composition-based stats.
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 86 GIVITASHNPKEYNGYKVYDENG---AQIIPPVDADIANA 122
G++ITASHN NG KV D +G +Q P IANA
Sbjct 62 GLMITASHNKVSDNGIKVSDPSGFMLSQEWEPFADQIANA 101
> dre:394000 pgm1, MGC63718, zgc:63718; phosphoglucomutase 1 (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=561
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 71 TPLVAFGVLHLNCVAGIVITASHNPKEYNG 100
TP V+ + + + GI++TASHNP NG
Sbjct 96 TPAVSCVIRKIKAIGGIILTASHNPGGPNG 125
> hsa:80164 hypothetical protein FLJ22184
Length=1346
Score = 31.6 bits (70), Expect = 0.86, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 84 VAGIVITASHNPKEYNGYKVYDENGAQIIPPVDADIA 120
+AG +A+ P+E GY E GA PP DA++A
Sbjct 1119 LAGHSSSATSTPEELRGYDSGPEGGAAASPPPDAELA 1155
> mmu:72157 Pgm2, 2610020G18Rik, AA407108, AI098105, Pgm-2, Pgm1;
phosphoglucomutase 2 (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=562
Score = 31.2 bits (69), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 69 VHTPLVAFGVLHLNCVAGIVITASHNPKEYNG 100
+ TP V+ + + + GI++TASHNP NG
Sbjct 94 LSTPAVSCIIRKIKAIGGIILTASHNPGGPNG 125
> sce:YMR105C PGM2, GAL5; Pgm2p (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=569
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 41/97 (42%), Gaps = 11/97 (11%)
Query 34 IIIGRDCRHNSERFSHLAAATFLSRGFRVYLC--RSAVHTPLVA--FGVLHLNCVAGIVI 89
+++G D R+ ++ H AA + G + + + TP + C GI++
Sbjct 57 LVVGGDGRYYNDVILHKIAAIGAANGIKKLVIGQHGLLSTPAASHIMRTYEEKCTGGIIL 116
Query 90 TASHN---PKEYNGYKVYDENGAQIIPPVDADIANAI 123
TASHN P+ G K NG P + NAI
Sbjct 117 TASHNPGGPENDMGIKYNLSNGG----PAPESVTNAI 149
> dre:560297 novel protein similar to vertebrate phosphoglucomutase
5 (PGM5)
Length=567
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 35/71 (49%), Gaps = 6/71 (8%)
Query 34 IIIGRDCRHNS----ERFSHLAAATFLSRGFRVYLCRSAVHTPLVAFGVLHLNCVAGIVI 89
+++G D R+ S E +AAA + R V + TP V+ + + + GI++
Sbjct 61 MVVGSDGRYFSRAATEVIVQMAAANGIGR--LVIGHNGILSTPAVSCIIRKIKAIGGIIL 118
Query 90 TASHNPKEYNG 100
TASH+P G
Sbjct 119 TASHSPGGPGG 129
> cpv:cgd2_3270 phosphoglucomutase, tandemly duplicated gene (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=670
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 23/43 (53%), Gaps = 7/43 (16%)
Query 84 VAGIVITASHNP---KEYNGYKVYDENGAQIIPPVDADIANAI 123
V GI++TASHNP E G K ++NG P + NAI
Sbjct 210 VGGILLTASHNPGGIDEDFGVKFNEKNGG----PAQDSVTNAI 248
> xla:379864 pgm1, MGC53760, pgm2; phosphoglucomutase 1 (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=562
Score = 28.9 bits (63), Expect = 5.1, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 71 TPLVAFGVLHLNCVAGIVITASHNPKEYNG 100
TP V+ + + GI++TASHNP NG
Sbjct 96 TPAVSCIIRKVKANGGIILTASHNPGGPNG 125
> ath:AT1G70820 phosphoglucomutase, putative / glucose phosphomutase,
putative (EC:5.4.2.8)
Length=615
Score = 28.5 bits (62), Expect = 6.4, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 85 AGIVITASHNPKEYNGYKVYDENGAQIIPPVD 116
A I++TASH P NG K + + G P V+
Sbjct 175 ASIMMTASHLPYTRNGLKFFTKRGGLTSPEVE 206
Lambda K H
0.322 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40