bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1441_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
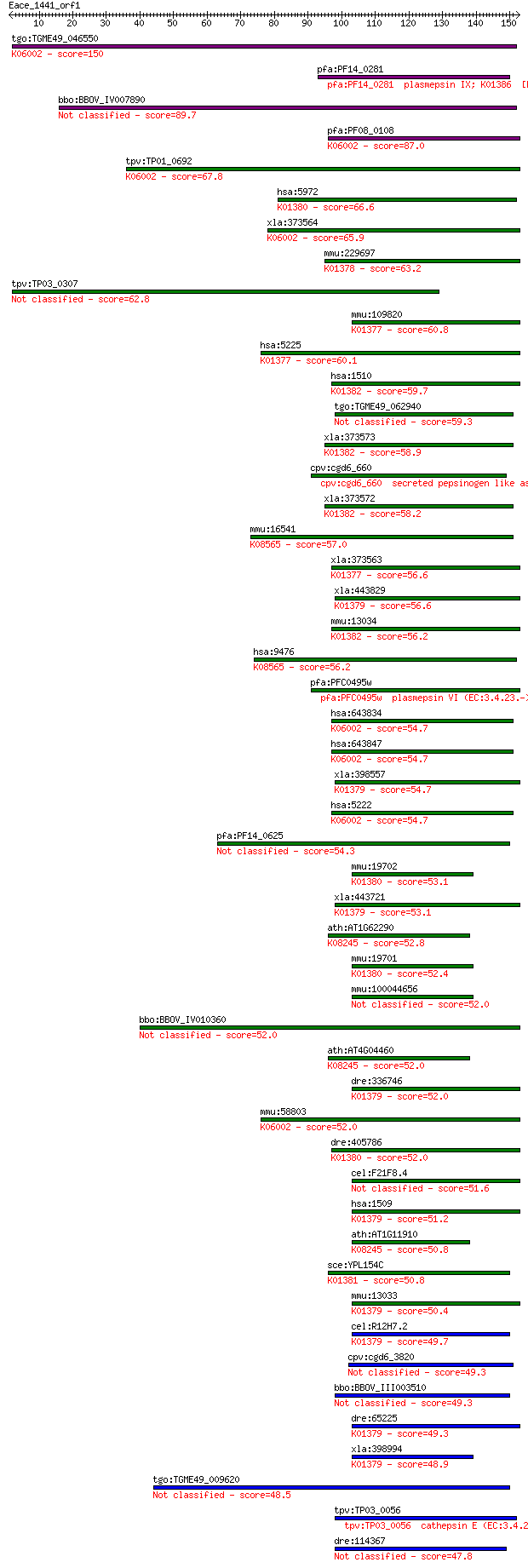
tgo:TGME49_046550 eukaryotic aspartyl protease, putative (EC:3... 150 1e-36
pfa:PF14_0281 plasmepsin IX; K01386 [EC:3.4.23.-] 90.1 3e-18
bbo:BBOV_IV007890 23.m06476; aspartyl protease 89.7 3e-18
pfa:PF08_0108 plasmepsin X (EC:3.4.23.1); K06002 pepsin A [EC:... 87.0 2e-17
tpv:TP01_0692 pepsinogen; K06002 pepsin A [EC:3.4.23.1] 67.8 1e-11
hsa:5972 REN, FLJ10761, HNFJ2; renin (EC:3.4.23.15); K01380 re... 66.6 3e-11
xla:373564 pga4, pga-A; pepsinogen 4, group I (pepsinogen A); ... 65.9 5e-11
mmu:229697 Cym, Gm131; chymosin (EC:3.4.23.4); K01378 chymosin... 63.2 3e-10
tpv:TP03_0307 hypothetical protein 62.8 5e-10
mmu:109820 Pgc, 2210410L06Rik, MGC117575, Upg-1, Upg1; progast... 60.8 1e-09
hsa:5225 PGC, FLJ99563, PEPC, PGII; progastricsin (pepsinogen ... 60.1 3e-09
hsa:1510 CTSE, CATE; cathepsin E (EC:3.4.23.34); K01382 cathep... 59.7 4e-09
tgo:TGME49_062940 eukaryotic aspartyl protease, putative (EC:3... 59.3 5e-09
xla:373573 ctse-b, CE2, cate, ce1, ce2-A; cathepsin E (EC:3.4.... 58.9 7e-09
cpv:cgd6_660 secreted pepsinogen like aspartyl protease having... 58.5 7e-09
xla:373572 ctse-a, CE1, cate, ce1-A, ce2, ctse; cathepsin E (E... 58.2 1e-08
mmu:16541 Napsa, KAP, Kdap, NAP1, SNAPA, pronapsin; napsin A a... 57.0 2e-08
xla:373563 pgc-A; pepsinogen C (EC:3.4.23.3); K01377 gastricsi... 56.6 3e-08
xla:443829 MGC82347 protein; K01379 cathepsin D [EC:3.4.23.5] 56.6 3e-08
mmu:13034 Ctse, A430072O03Rik, C920004C08Rik, CE, CatE; cathep... 56.2 4e-08
hsa:9476 NAPSA, KAP, Kdap, NAP1, NAPA, SNAPA; napsin A asparti... 56.2 4e-08
pfa:PFC0495w plasmepsin VI (EC:3.4.23.-); K01386 [EC:3.4.23.-] 55.1 8e-08
hsa:643834 PGA3, DKFZp666J2410, PGA4, PGA5; pepsinogen 3, grou... 54.7 1e-07
hsa:643847 PGA4, FLJ58952, FLJ77962, PGA3, PGA5; pepsinogen 4,... 54.7 1e-07
xla:398557 cathepsin D (EC:3.4.23.5); K01379 cathepsin D [EC:3... 54.7 1e-07
hsa:5222 PGA5, PGA3, PGA4; pepsinogen 5, group I (pepsinogen A... 54.7 1e-07
pfa:PF14_0625 plasmepsin VIII (EC:3.4.17.4) 54.3 1e-07
mmu:19702 Ren2, Ren, Ren-2, Ren-B, Rn-2, Rnr; renin 2 tandem d... 53.1 4e-07
xla:443721 ctsd; cathepsin D (EC:3.4.23.5); K01379 cathepsin D... 53.1 4e-07
ath:AT1G62290 aspartyl protease family protein; K08245 phyteps... 52.8 4e-07
mmu:19701 Ren1, D19352, Ren, Ren-1, Ren-A, Ren1c, Ren1d, Rn-1,... 52.4 6e-07
mmu:100044656 renin-1-like 52.0 7e-07
bbo:BBOV_IV010360 23.m06120; aspartyl protease 52.0 7e-07
ath:AT4G04460 aspartyl protease family protein; K08245 phyteps... 52.0 7e-07
dre:336746 fa94d12, wu:fa94d12; zgc:63831; K01379 cathepsin D ... 52.0 7e-07
mmu:58803 Pga5, 1110035E17Rik, Pepf; pepsinogen 5, group I (EC... 52.0 7e-07
dre:405786 ren; renin (EC:3.4.23.15); K01380 renin [EC:3.4.23.15] 52.0 8e-07
cel:F21F8.4 hypothetical protein 51.6 9e-07
hsa:1509 CTSD, CLN10, CPSD, MGC2311; cathepsin D (EC:3.4.23.5)... 51.2 1e-06
ath:AT1G11910 aspartyl protease family protein; K08245 phyteps... 50.8 1e-06
sce:YPL154C PEP4, PHO9, PRA1, yscA; Pep4p (EC:3.4.23.25); K013... 50.8 2e-06
mmu:13033 Ctsd, CD, CatD; cathepsin D (EC:3.4.23.5); K01379 ca... 50.4 2e-06
cel:R12H7.2 asp-4; ASpartyl Protease family member (asp-4); K0... 49.7 4e-06
cpv:cgd6_3820 membrane bound aspartyl proteinase with a signal... 49.3 4e-06
bbo:BBOV_III003510 17.m07331; eukaryotic aspartyl protease fam... 49.3 4e-06
dre:65225 ctsd, catD, fb93e11, fj17b09, wu:fb93e11, wu:fj17b09... 49.3 5e-06
xla:398994 napsa, MGC68767, kap, kdap, nap1, napa, snapa; naps... 48.9 7e-06
tgo:TGME49_009620 eukaryotic aspartyl protease, putative (EC:3... 48.5 7e-06
tpv:TP03_0056 cathepsin E (EC:3.4.23.34); K01386 [EC:3.4.23.-] 48.5 8e-06
dre:114367 nots, LAP, MGC192423, ctsel, zgc:103608; nothepsin 47.8 1e-05
> tgo:TGME49_046550 eukaryotic aspartyl protease, putative (EC:3.4.23.1);
K06002 pepsin A [EC:3.4.23.1]
Length=643
Score = 150 bits (380), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 77/171 (45%), Positives = 103/171 (60%), Gaps = 21/171 (12%)
Query 2 MPMTRAVSLIDGEDSAFAALEKNHSAFLNTRTAFFMKRGEGPFGALGNKLGQTSAAAAAA 61
MPM ++ + + LE NH LN + F++ RG+GPFG+LGN G AA A
Sbjct 156 MPMMPIRNIDTHREETLSRLEANHRTHLNEKKNFYVLRGKGPFGSLGNLPGTPGLDAAIA 215
Query 62 ADAAAADAPAAPLSFLQHP---------------------AQPAATLELGLAQTSVPILQ 100
++ D P+A + +++ L+L LA+TSVPILQ
Sbjct 216 FGLSSPDLPSASFAQIKNKDSSDSGDVAAVGEETDSAVADGSKTLDLDLKLAETSVPILQ 275
Query 101 LKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPS 151
+KDSQ+ G++GIGTPPQ+V+PIFDTGSTNLWVV S+CTD+TC KVTRFDPS
Sbjct 276 MKDSQYVGVIGIGTPPQFVQPIFDTGSTNLWVVGSKCTDDTCTKVTRFDPS 326
> pfa:PF14_0281 plasmepsin IX; K01386 [EC:3.4.23.-]
Length=627
Score = 90.1 bits (222), Expect = 3e-18, Method: Composition-based stats.
Identities = 34/57 (59%), Positives = 49/57 (85%), Gaps = 0/57 (0%)
Query 93 QTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
+ ++P+ QL+DSQ+ G + IGTPPQ +RPIFDTGSTN+W+VS++C DETC+KV R++
Sbjct 215 KVTLPLQQLEDSQYVGYIQIGTPPQTIRPIFDTGSTNIWIVSTKCKDETCLKVHRYN 271
> bbo:BBOV_IV007890 23.m06476; aspartyl protease
Length=463
Score = 89.7 bits (221), Expect = 3e-18, Method: Composition-based stats.
Identities = 52/138 (37%), Positives = 70/138 (50%), Gaps = 19/138 (13%)
Query 16 SAFAALEKNHSAFLNTRTAFFMKRGEGPFGALGNKLGQTSAAAAAAADAAAADAPAAPLS 75
SA E NH+AF + +F +R EG FG++G S A
Sbjct 76 SALQMAEDNHNAFRRGKISF-ARRSEGSFGSIGYSGSHVSQNALK--------------- 119
Query 76 FLQHPAQPAATLELGLAQTS--VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVV 133
L + P + L ++ +PI + KDS + G + IGTPPQ V PIFDTGSTNLWVV
Sbjct 120 -LMRRSNPKSLSMLSRSEYGKYIPIKETKDSLYIGEIMIGTPPQIVHPIFDTGSTNLWVV 178
Query 134 SSRCTDETCVKVTRFDPS 151
C D +C+KV R++ S
Sbjct 179 GYDCKDPSCLKVARYNTS 196
> pfa:PF08_0108 plasmepsin X (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=573
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 36/57 (63%), Positives = 46/57 (80%), Gaps = 0/57 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
VP+ L+DSQF G L +GTPPQ V PIFDTGSTN+WVV++ C +E+C KV R+DP+K
Sbjct 238 VPLKHLRDSQFVGELLVGTPPQTVYPIFDTGSTNVWVVTTACEEESCKKVRRYDPNK 294
> tpv:TP01_0692 pepsinogen; K06002 pepsin A [EC:3.4.23.1]
Length=377
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 59/127 (46%), Gaps = 21/127 (16%)
Query 36 FMKRGEGPFGALG-NKLGQTSAAAAAAADAAAADAPAAPLS---------FLQHPAQPAA 85
F+ R +GP+G+LG ++ + + ++ D ++ +HP +
Sbjct 71 FLYRSQGPYGSLGTSQHNSHNCNSYHTHNSVNTDDSVNSVNSVDGTRLTPIREHPMKKC- 129
Query 86 TLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKV 145
+PI L+ Q+ +G+GTP Q + PI DTGSTN WV+S +C TC V
Sbjct 130 ----------IPIPHLRHVQYVMSIGVGTPKQEIYPIIDTGSTNTWVISEQCNSITCSGV 179
Query 146 TRFDPSK 152
F+ K
Sbjct 180 PTFNSRK 186
> hsa:5972 REN, FLJ10761, HNFJ2; renin (EC:3.4.23.15); K01380
renin [EC:3.4.23.15]
Length=406
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 81 AQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTD- 139
+QP L LG +SV + D+Q+ G +GIGTPPQ + +FDTGS+N+WV SS+C+
Sbjct 61 SQPMKRLTLGNTTSSVILTNYMDTQYYGEIGIGTPPQTFKVVFDTGSSNVWVPSSKCSRL 120
Query 140 -ETCVKVTRFDPS 151
CV FD S
Sbjct 121 YTACVYHKLFDAS 133
> xla:373564 pga4, pga-A; pepsinogen 4, group I (pepsinogen A);
K06002 pepsin A [EC:3.4.23.1]
Length=384
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 44/76 (57%), Gaps = 1/76 (1%)
Query 78 QHPAQPAATLELGLAQTSVPILQ-LKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSR 136
++P PA+ LAQ+S LQ D ++ G + IGTPPQ IFDTGS NLWV S
Sbjct 43 KNPYNPASKYFPTLAQSSAETLQNYMDIEYYGTISIGTPPQEFTVIFDTGSANLWVPSVY 102
Query 137 CTDETCVKVTRFDPSK 152
C+ + C RF+P +
Sbjct 103 CSSQACSNHNRFNPQQ 118
> mmu:229697 Cym, Gm131; chymosin (EC:3.4.23.4); K01378 chymosin
[EC:3.4.23.4]
Length=379
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 36/58 (62%), Gaps = 0/58 (0%)
Query 95 SVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
S P++ DS++ G + IGTPPQ +FDTGS+ LWV S C + C RFDPSK
Sbjct 63 SEPLINYLDSEYFGTIYIGTPPQEFTVVFDTGSSELWVPSVYCNSKVCRNHHRFDPSK 120
> tpv:TP03_0307 hypothetical protein
Length=159
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/134 (29%), Positives = 62/134 (46%), Gaps = 15/134 (11%)
Query 2 MPMTRAVSLIDGEDSAFAALE---KNHSAFLNTRTAFFMKRGEGPFGALGNKLG--QTSA 56
MP + L E + +E +N F N + +F + R +GPFG+LG+ + T+
Sbjct 29 MPKFMTIGLRRYEKERVSVIELARENQKNFKNGKISFVL-RSQGPFGSLGHTITNPHTNT 87
Query 57 AAAAAADAAAADAPAAPLSFLQHPAQPAATLELGLAQ--TSVPILQLKDSQFCGMLGIGT 114
+ QH + + + + VP+ Q+KDS + G + +GT
Sbjct 88 HTNDLTNTLTNSQT-------QHSVRRKFNRNMNILKGYEYVPLQQIKDSLYVGTISVGT 140
Query 115 PPQWVRPIFDTGST 128
PPQ + PIFDTGST
Sbjct 141 PPQILHPIFDTGST 154
> mmu:109820 Pgc, 2210410L06Rik, MGC117575, Upg-1, Upg1; progastricsin
(pepsinogen C) (EC:3.4.23.3); K01377 gastricsin [EC:3.4.23.3]
Length=392
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/50 (52%), Positives = 33/50 (66%), Gaps = 0/50 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
D+ + G + IGTPPQ +FDTGS+NLWV S C E C TR++PSK
Sbjct 73 DASYYGEISIGTPPQNFLVLFDTGSSNLWVSSVYCQSEACTTHTRYNPSK 122
> hsa:5225 PGC, FLJ99563, PEPC, PGII; progastricsin (pepsinogen
C) (EC:3.4.23.3); K01377 gastricsin [EC:3.4.23.3]
Length=315
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 76 FLQ-HPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVS 134
FL+ H PA G + + D+ + G + IGTPPQ +FDTGS+NLWV S
Sbjct 42 FLRTHKYDPAWKYRFGDLSVTYEPMAYMDAAYFGEISIGTPPQNFLVLFDTGSSNLWVPS 101
Query 135 SRCTDETCVKVTRFDPSK 152
C + C +RF+PS+
Sbjct 102 VYCQSQACTSHSRFNPSE 119
> hsa:1510 CTSE, CATE; cathepsin E (EC:3.4.23.34); K01382 cathepsin
E [EC:3.4.23.34]
Length=396
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 35/56 (62%), Gaps = 0/56 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
P++ D ++ G + IG+PPQ IFDTGS+NLWV S CT C +RF PS+
Sbjct 69 PLINYLDMEYFGTISIGSPPQNFTVIFDTGSSNLWVPSVYCTSPACKTHSRFQPSQ 124
> tgo:TGME49_062940 eukaryotic aspartyl protease, putative (EC:3.4.23.34)
Length=469
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 25/53 (47%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
+L +SQ+ G + +GTPP +FDTGS+NLW+ +S C CV TRFDP
Sbjct 96 LLNFHNSQYFGEIQVGTPPVSFIVVFDTGSSNLWIPASECKQGGCVPHTRFDP 148
> xla:373573 ctse-b, CE2, cate, ce1, ce2-A; cathepsin E (EC:3.4.23.34);
K01382 cathepsin E [EC:3.4.23.34]
Length=397
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 95 SVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
S P++ D Q+ G + IGTPPQ IFDTGS+NLWV S C C + RF P
Sbjct 63 SEPLINYMDVQYFGEISIGTPPQNFTVIFDTGSSNLWVPSVYCISPACAQHNRFQP 118
> cpv:cgd6_660 secreted pepsinogen like aspartyl protease having
a signal peptide ; K01386 [EC:3.4.23.-]
Length=633
Score = 58.5 bits (140), Expect = 7e-09, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRF 148
+ ++P+ ++K+S F + IG P Q PI DTGS+NLWV+ C +C KV R+
Sbjct 176 ITNITIPLFEVKNSLFVCRIRIGEPEQEFWPIIDTGSSNLWVIGEECNQSSCQKVKRY 233
> xla:373572 ctse-a, CE1, cate, ce1-A, ce2, ctse; cathepsin E
(EC:3.4.23.34); K01382 cathepsin E [EC:3.4.23.34]
Length=397
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 95 SVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
S P++ D ++ G + +GTPPQ IFDTGS+NLWV S C + C + RF P
Sbjct 63 SEPLINYMDVEYFGEISVGTPPQNFTVIFDTGSSNLWVPSVYCISQACAQHDRFQP 118
> mmu:16541 Napsa, KAP, Kdap, NAP1, SNAPA, pronapsin; napsin A
aspartic peptidase; K08565 napsin-A [EC:3.4.23.-]
Length=419
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 47/80 (58%), Gaps = 2/80 (2%)
Query 73 PLSFLQHPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWV 132
PL+ + A+ + T G + VP+ + ++Q+ G +G+GTPPQ +FDTGS+NLWV
Sbjct 40 PLNGWEQLAELSRTSTSGGNPSFVPLSKFMNTQYFGTIGLGTPPQNFTVVFDTGSSNLWV 99
Query 133 VSSRCT--DETCVKVTRFDP 150
S+RC C RF+P
Sbjct 100 PSTRCHFFSLACWFHHRFNP 119
> xla:373563 pgc-A; pepsinogen C (EC:3.4.23.3); K01377 gastricsin
[EC:3.4.23.3]
Length=383
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
P+ D + G + IGTPPQ +FDTGS+NLWV S+ C + C F+PS+
Sbjct 58 PLSNYMDMSYYGEISIGTPPQNFLVLFDTGSSNLWVASTYCQSQACTNHPLFNPSQ 113
> xla:443829 MGC82347 protein; K01379 cathepsin D [EC:3.4.23.5]
Length=401
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 36/57 (63%), Gaps = 2/57 (3%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
++ D+Q+ G +GIGTPPQ +FDTGS+NLWV S C+ D C ++D SK
Sbjct 73 LMNYLDAQYYGEIGIGTPPQTFTVVFDTGSSNLWVPSVHCSMFDIACWMHHKYDSSK 129
> mmu:13034 Ctse, A430072O03Rik, C920004C08Rik, CE, CatE; cathepsin
E (EC:3.4.23.34); K01382 cathepsin E [EC:3.4.23.34]
Length=397
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
P++ D ++ G + IGTPPQ IFDTGS+NLWV S CT C F PS+
Sbjct 70 PLINYLDMEYFGTISIGTPPQNFTVIFDTGSSNLWVPSVYCTSPACKAHPVFHPSQ 125
> hsa:9476 NAPSA, KAP, Kdap, NAP1, NAPA, SNAPA; napsin A aspartic
peptidase; K08565 napsin-A [EC:3.4.23.-]
Length=420
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 44/85 (51%), Gaps = 7/85 (8%)
Query 74 LSFLQHPAQPAATLELGLAQTS-----VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGST 128
L+ L+ +PA +LG VP+ +D Q+ G +G+GTPPQ FDTGS+
Sbjct 41 LNLLRGWREPAELPKLGAPSPGDKPIFVPLSNYRDVQYFGEIGLGTPPQNFTVAFDTGSS 100
Query 129 NLWVVSSRCT--DETCVKVTRFDPS 151
NLWV S RC C RFDP
Sbjct 101 NLWVPSRRCHFFSVPCWLHHRFDPK 125
> pfa:PFC0495w plasmepsin VI (EC:3.4.23.-); K01386 [EC:3.4.23.-]
Length=432
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 37/62 (59%), Gaps = 0/62 (0%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
L+ +L +SQF +G+G PPQ + +FDTGS+NL + S++C C +F+P
Sbjct 85 LSYIQEDLLNFHNSQFIADIGVGNPPQVFKVVFDTGSSNLAIPSTKCIKGGCASHKKFNP 144
Query 151 SK 152
+K
Sbjct 145 NK 146
> hsa:643834 PGA3, DKFZp666J2410, PGA4, PGA5; pepsinogen 3, group
I (pepsinogen A) (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=388
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
P+ D ++ G +GIGTP Q +FDTGS+NLWV S C+ C RF+P
Sbjct 67 PLENYLDMEYFGTIGIGTPAQDFTVVFDTGSSNLWVPSVYCSSLACTNHNRFNP 120
> hsa:643847 PGA4, FLJ58952, FLJ77962, PGA3, PGA5; pepsinogen
4, group I (pepsinogen A) (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=388
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
P+ D ++ G +GIGTP Q +FDTGS+NLWV S C+ C RF+P
Sbjct 67 PLENYLDMEYFGTIGIGTPAQDFTVVFDTGSSNLWVPSVYCSSLACTNHNRFNP 120
> xla:398557 cathepsin D (EC:3.4.23.5); K01379 cathepsin D [EC:3.4.23.5]
Length=409
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 35/57 (61%), Gaps = 2/57 (3%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
+L D+Q+ G + IGTPPQ +FDTGS+NLWV S C+ D C ++D SK
Sbjct 71 LLNYLDAQYYGEISIGTPPQPFTVVFDTGSSNLWVASVHCSMFDIACWMHRKYDSSK 127
> hsa:5222 PGA5, PGA3, PGA4; pepsinogen 5, group I (pepsinogen
A) (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=388
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
P+ D ++ G +GIGTP Q +FDTGS+NLWV S C+ C RF+P
Sbjct 67 PLENYLDMEYFGTIGIGTPAQDFTVVFDTGSSNLWVPSVYCSSLACTNHNRFNP 120
> pfa:PF14_0625 plasmepsin VIII (EC:3.4.17.4)
Length=385
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 41/91 (45%), Gaps = 4/91 (4%)
Query 63 DAAAADAPAAPLSFLQHPAQPAATLELGLAQTSVPILQLK----DSQFCGMLGIGTPPQW 118
+ A A L ++ A L L + + LK + QF G + IG PPQ
Sbjct 15 NIIAVKAFKENLRVSKYYAGGKHKLNLENKYIGISTIVLKGGYINRQFIGEINIGNPPQT 74
Query 119 VRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
+ +FDTGSTNLW+ S C C ++D
Sbjct 75 FKVLFDTGSTNLWIPSKNCFTRACYNKRKYD 105
> mmu:19702 Ren2, Ren, Ren-2, Ren-B, Rn-2, Rnr; renin 2 tandem
duplication of Ren1 (EC:3.4.23.15); K01380 renin [EC:3.4.23.15]
Length=424
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT 138
+SQ+ G +GIGTPPQ + IFDTGS NLWV S++C+
Sbjct 103 NSQYYGEIGIGTPPQTFKVIFDTGSANLWVPSTKCS 138
> xla:443721 ctsd; cathepsin D (EC:3.4.23.5); K01379 cathepsin
D [EC:3.4.23.5]
Length=399
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 35/57 (61%), Gaps = 2/57 (3%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
+L D+Q+ G + IGTPPQ +FDTGS+NLWV S C+ D C ++D SK
Sbjct 71 LLNYLDAQYYGEISIGTPPQPFTVVFDTGSSNLWVPSVHCSFWDIACWLHHKYDSSK 127
> ath:AT1G62290 aspartyl protease family protein; K08245 phytepsin
[EC:3.4.23.40]
Length=513
Score = 52.8 bits (125), Expect = 4e-07, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 29/42 (69%), Gaps = 0/42 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
VP+ D+Q+ G + IGTPPQ IFDTGS+NLWV S +C
Sbjct 79 VPLKNYLDAQYYGEIAIGTPPQKFTVIFDTGSSNLWVPSGKC 120
> mmu:19701 Ren1, D19352, Ren, Ren-1, Ren-A, Ren1c, Ren1d, Rn-1,
Rnr; renin 1 structural (EC:3.4.23.15); K01380 renin [EC:3.4.23.15]
Length=402
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT 138
++Q+ G +GIGTPPQ + IFDTGS NLWV S++C+
Sbjct 81 NTQYYGEIGIGTPPQTFKVIFDTGSANLWVPSTKCS 116
> mmu:100044656 renin-1-like
Length=425
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT 138
++Q+ G +GIGTPPQ + IFDTGS NLWV S++C+
Sbjct 104 NTQYYGEIGIGTPPQTFKVIFDTGSANLWVPSTKCS 139
> bbo:BBOV_IV010360 23.m06120; aspartyl protease
Length=532
Score = 52.0 bits (123), Expect = 7e-07, Method: Composition-based stats.
Identities = 34/124 (27%), Positives = 54/124 (43%), Gaps = 16/124 (12%)
Query 40 GEGPFGALG----------NKLGQTSAAAAAAADAAAADAPAAPLSFL-QHPAQPAATLE 88
G GP G++G ++ + S AA D ++ P++ Q A P
Sbjct 113 GRGPNGSMGVDMETKRRHEERIRKQSLEQLAAMDNNTSEKDEIPVTVTRQETAFPRVP-- 170
Query 89 LGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRF 148
A + L+ + F IG P Q PI DTGSTN+W + CT C++ ++
Sbjct 171 ---ANACLDFASLRHTTFVVEAYIGKPGQKFLPILDTGSTNVWAIHPECTSAGCMEAIKY 227
Query 149 DPSK 152
DP++
Sbjct 228 DPAR 231
> ath:AT4G04460 aspartyl protease family protein; K08245 phytepsin
[EC:3.4.23.40]
Length=508
Score = 52.0 bits (123), Expect = 7e-07, Method: Composition-based stats.
Identities = 22/42 (52%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
VP+ D+Q+ G + IGTPPQ IFDTGS+NLW+ S++C
Sbjct 77 VPLKNYLDAQYYGDITIGTPPQKFTVIFDTGSSNLWIPSTKC 118
> dre:336746 fa94d12, wu:fa94d12; zgc:63831; K01379 cathepsin
D [EC:3.4.23.5]
Length=412
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 25/52 (48%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
D+Q+ GM+ IGTPPQ +FDTGS+NLWV S C D C R++ K
Sbjct 89 DAQYYGMISIGTPPQDFSVLFDTGSSNLWVPSIHCAFLDIACWLHRRYNSKK 140
> mmu:58803 Pga5, 1110035E17Rik, Pepf; pepsinogen 5, group I (EC:3.4.23.1);
K06002 pepsin A [EC:3.4.23.1]
Length=387
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 37/77 (48%), Gaps = 8/77 (10%)
Query 76 FLQHPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSS 135
L+ PA T E P+ D + G++ IGTPPQ R + DTGS+ LWV S
Sbjct 52 LLEQRRNPAVTYE--------PMRNYLDLVYIGIISIGTPPQEFRVVLDTGSSVLWVPSI 103
Query 136 RCTDETCVKVTRFDPSK 152
C+ C F+P +
Sbjct 104 YCSSPACAHHKAFNPLR 120
> dre:405786 ren; renin (EC:3.4.23.15); K01380 renin [EC:3.4.23.15]
Length=395
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
P++ D+Q+ G + IG+P Q +FDTGS NLWV S C+ C R+D SK
Sbjct 67 PLINYLDTQYFGEISIGSPAQMFNVVFDTGSANLWVPSHSCSPLYTACFTHNRYDASK 124
> cel:F21F8.4 hypothetical protein
Length=395
Score = 51.6 bits (122), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 25/50 (50%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
D ++ G + IGTPPQ + DTGS NLWV S C D TC + FD SK
Sbjct 70 DVEYLGNITIGTPPQQFIVVLDTGSANLWVPGSNC-DGTCKGKSEFDSSK 118
> hsa:1509 CTSD, CLN10, CPSD, MGC2311; cathepsin D (EC:3.4.23.5);
K01379 cathepsin D [EC:3.4.23.5]
Length=412
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
D+Q+ G +GIGTPPQ +FDTGS+NLWV S C D C +++ K
Sbjct 76 DAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIHHKYNSDK 127
> ath:AT1G11910 aspartyl protease family protein; K08245 phytepsin
[EC:3.4.23.40]
Length=506
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
D+Q+ G + IGTPPQ +FDTGS+NLWV SS+C
Sbjct 79 DAQYYGEIAIGTPPQKFTVVFDTGSSNLWVPSSKC 113
> sce:YPL154C PEP4, PHO9, PRA1, yscA; Pep4p (EC:3.4.23.25); K01381
saccharopepsin [EC:3.4.23.25]
Length=405
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
VP+ ++Q+ + +GTPPQ + I DTGS+NLWV S+ C C +++D
Sbjct 81 VPLTNYLNAQYYTDITLGTPPQNFKVILDTGSSNLWVPSNECGSLACFLHSKYD 134
> mmu:13033 Ctsd, CD, CatD; cathepsin D (EC:3.4.23.5); K01379
cathepsin D [EC:3.4.23.5]
Length=410
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
D+Q+ G +GIGTPPQ +FDTGS+NLWV S C D C +++ K
Sbjct 76 DAQYYGDIGIGTPPQCFTVVFDTGSSNLWVPSIHCKILDIACWVHHKYNSDK 127
> cel:R12H7.2 asp-4; ASpartyl Protease family member (asp-4);
K01379 cathepsin D [EC:3.4.23.5]
Length=444
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 31/49 (63%), Gaps = 2/49 (4%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFD 149
D+Q+ G + IGTP Q IFDTGS+NLW+ S +C D C+ R+D
Sbjct 91 DAQYFGTISIGTPAQNFTVIFDTGSSNLWIPSKKCPFYDIACMLHHRYD 139
> cpv:cgd6_3820 membrane bound aspartyl proteinase with a signal
peptide plus transmembrane domain
Length=467
Score = 49.3 bits (116), Expect = 4e-06, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 32/49 (65%), Gaps = 0/49 (0%)
Query 102 KDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
++SQ+ G + +GTPP+ IFDTGS+++W+ S C + C ++DP
Sbjct 92 QNSQYFGKIEVGTPPREFVVIFDTGSSSVWIPSIECKHKGCEPHNKYDP 140
> bbo:BBOV_III003510 17.m07331; eukaryotic aspartyl protease family
protein
Length=521
Score = 49.3 bits (116), Expect = 4e-06, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 32/52 (61%), Gaps = 0/52 (0%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
+L +++Q+ G + +GTPP+ +FDTGS+ LW+ S C+ C +FD
Sbjct 141 LLNFENNQYFGEIEVGTPPEKFVVVFDTGSSQLWIPSKECSSTGCSTHRKFD 192
> dre:65225 ctsd, catD, fb93e11, fj17b09, wu:fb93e11, wu:fj17b09;
cathepsin D (EC:3.4.23.5); K01379 cathepsin D [EC:3.4.23.5]
Length=399
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 33/52 (63%), Gaps = 2/52 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFDPSK 152
D+Q+ G +G+GTP Q +FDTGS+NLWV S C TD C+ +++ K
Sbjct 72 DAQYYGEIGLGTPVQTFTVVFDTGSSNLWVPSVHCSLTDIACLLHHKYNGGK 123
> xla:398994 napsa, MGC68767, kap, kdap, nap1, napa, snapa; napsin
A aspartic peptidase; K01379 cathepsin D [EC:3.4.23.5]
Length=392
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 27/36 (75%), Gaps = 0/36 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT 138
D+Q+ G + IGTPPQ IFDTGS+NLWV S +C+
Sbjct 64 DAQYYGEIFIGTPPQKFAVIFDTGSSNLWVPSVKCS 99
> tgo:TGME49_009620 eukaryotic aspartyl protease, putative (EC:3.4.23.34)
Length=432
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 49/106 (46%), Gaps = 6/106 (5%)
Query 44 FGALGNKLGQTSAAAAAAADAAAADAPAAPLSFLQHPAQPAATLELGLAQTSVPILQLKD 103
F L L S+ + A+ A A A L ++ P + L V + ++
Sbjct 13 FPLLWAVLTYESSWSVASPTAPGELAEAKLLPLIKRHDVPHTRVLL------VSLQNHRN 66
Query 104 SQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
+Q+ G + +G PPQ +FDTGS + W+ S+ C +C +RFD
Sbjct 67 TQYFGKISVGNPPQSFNVVFDTGSHHFWIPSNECQASSCRAHSRFD 112
> tpv:TP03_0056 cathepsin E (EC:3.4.23.34); K01386 [EC:3.4.23.-]
Length=513
Score = 48.5 bits (114), Expect = 8e-06, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDET---CVKVTRFDPS 151
+L ++SQ+ G + +GTPP+ +FDTGS+ LW+ S C + + C + FD S
Sbjct 153 LLNFENSQYFGEIQVGTPPKSFVVVFDTGSSQLWIPSKACLNHSSNGCARHRMFDSS 209
> dre:114367 nots, LAP, MGC192423, ctsel, zgc:103608; nothepsin
Length=416
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 98 ILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRF 148
+ D+QF G + +G P Q +FDTGS++LWV SS C + C +F
Sbjct 78 LYNFMDAQFFGQISLGRPEQNFTVVFDTGSSDLWVPSSYCVTQACALHNKF 128
Lambda K H
0.318 0.130 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40