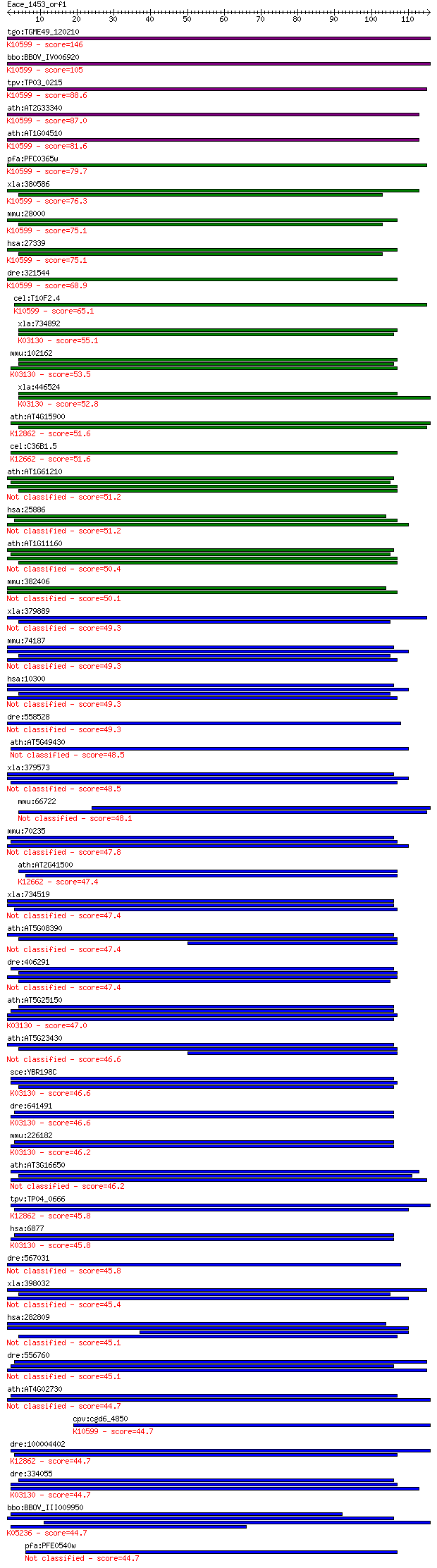
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1453_orf1
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_120210 WD-40 repeat protein, putative ; K10599 pre-... 146 1e-35
bbo:BBOV_IV006920 23.m06216; WD domain, G-beta repeat containi... 105 4e-23
tpv:TP03_0215 hypothetical protein; K10599 pre-mRNA-processing... 88.6 4e-18
ath:AT2G33340 nucleotide binding / ubiquitin-protein ligase; K... 87.0 1e-17
ath:AT1G04510 transducin family protein / WD-40 repeat family ... 81.6 5e-16
pfa:PFC0365w PRP19-like protein, putative; K10599 pre-mRNA-pro... 79.7 2e-15
xla:380586 prpf19, MGC52755, nmp200; PRP19/PSO4 pre-mRNA proce... 76.3 3e-14
mmu:28000 Prpf19, AA617263, AL024362, D19Wsu55e, NMP200, PSO4,... 75.1 5e-14
hsa:27339 PRPF19, NMP200, PRP19, PSO4, SNEV, UBOX4, hPSO4; PRP... 75.1 5e-14
dre:321544 prp19, NMP200, fb18f09, wu:fb18f09, zgc:56158; PRP1... 68.9 4e-12
cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing f... 65.1 5e-11
xla:734892 hypothetical protein MGC130867; K03130 transcriptio... 55.1 6e-08
mmu:102162 Taf5l, 1110005N04Rik, AI849020; TAF5-like RNA polym... 53.5 2e-07
xla:446524 taf5l, MGC80243; TAF5-like RNA polymerase II, p300/... 52.8 2e-07
ath:AT4G15900 PRL1; PRL1 (PLEIOTROPIC REGULATORY LOCUS 1); bas... 51.6 5e-07
cel:C36B1.5 prp-4; yeast PRP (splicing factor) related family ... 51.6 6e-07
ath:AT1G61210 WD-40 repeat family protein / katanin p80 subuni... 51.2 8e-07
hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1 c... 51.2 8e-07
ath:AT1G11160 nucleotide binding 50.4 1e-06
mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar prote... 50.1 2e-06
xla:379889 katnb1, MGC52971; katanin p80 (WD40-containing) sub... 49.3 3e-06
mmu:74187 Katnb1, 2410003J24Rik, KAT; katanin p80 (WD40-contai... 49.3 3e-06
hsa:10300 KATNB1, KAT; katanin p80 (WD repeat containing) subu... 49.3 3e-06
dre:558528 wdr47, si:ch211-66k16.12; WD repeat domain 47 49.3
ath:AT5G49430 transducin family protein / WD-40 repeat family ... 48.5 5e-06
xla:379573 poc1a, MGC69111, pix2, wdr51a; POC1 centriolar prot... 48.5 5e-06
mmu:66722 Spag16, 4921511D23Rik, 4930524F24Rik, 4930585K05Rik,... 48.1 6e-06
mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protei... 47.8 9e-06
ath:AT2G41500 EMB2776; nucleotide binding; K12662 U4/U6 small ... 47.4 1e-05
xla:734519 poc1b, MGC114911, pix1, tuwd12, wdr51b; POC1 centri... 47.4 1e-05
ath:AT5G08390 hypothetical protein 47.4 1e-05
dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin ... 47.4 1e-05
ath:AT5G25150 TAF5; TAF5 (TBP-ASSOCIATED FACTOR 5); nucleotide... 47.0 1e-05
ath:AT5G23430 transducin family protein / WD-40 repeat family ... 46.6 2e-05
sce:YBR198C TAF5, TAF90; Taf5p; K03130 transcription initiatio... 46.6 2e-05
dre:641491 taf5, MGC123129, wu:fc38g05, zgc:123129; TAF5 RNA p... 46.6 2e-05
mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase ... 46.2 2e-05
ath:AT3G16650 PP1/PP2A phosphatases pleiotropic regulator 2 (P... 46.2 3e-05
tpv:TP04_0666 hypothetical protein; K12862 pleiotropic regulat... 45.8 3e-05
hsa:6877 TAF5, TAF2D, TAFII100; TAF5 RNA polymerase II, TATA b... 45.8 3e-05
dre:567031 WD repeat-containing protein 47-like 45.8 4e-05
xla:398032 katnb1; katanin p80 (WD repeat containing) subunit B 1 45.4
hsa:282809 POC1B, FLJ14923, FLJ41111, PIX1, TUWD12, WDR51B; PO... 45.1 6e-05
dre:556760 wdr69, si:dkey-223n17.5, zgc:153626, zgc:153780; WD... 45.1 6e-05
ath:AT4G02730 transducin family protein / WD-40 repeat family ... 44.7 7e-05
cpv:cgd6_4850 PRP19 non-snRNP sliceosome component required fo... 44.7 7e-05
dre:100004402 plrg1, MGC174842, MGC55651, MGC86603, wu:fi74f01... 44.7 7e-05
dre:334055 taf5l, fi28g02, wu:fi28g02, zgc:63765; TAF5-like RN... 44.7 7e-05
bbo:BBOV_III009950 17.m07862; WD domain, G-beta repeat domain ... 44.7 8e-05
pfa:PFE0540w WD-repeat protein, putative 44.7 8e-05
> tgo:TGME49_120210 WD-40 repeat protein, putative ; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=515
Score = 146 bits (369), Expect = 1e-35, Method: Composition-based stats.
Identities = 64/115 (55%), Positives = 86/115 (74%), Gaps = 2/115 (1%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
+V+S SDDKTV+IWR S D S + +R+ RG++ L+LHPL +YFA+ AAD TW+F
Sbjct 268 VVVSASDDKTVRIWRASGD--SPYKTAATIRKHRGEVTCLSLHPLGNYFASCAADKTWAF 325
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAALQ 115
I+EGR L + K+L QY ++FHPDGMI+GGGGVDG+++IWDMKG RAAL+
Sbjct 326 SDIQEGRCLQMQKNLPCQYKCVSFHPDGMILGGGGVDGSVHIWDMKGLAYRAALK 380
> bbo:BBOV_IV006920 23.m06216; WD domain, G-beta repeat containing
protein; K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=495
Score = 105 bits (262), Expect = 4e-23, Method: Composition-based stats.
Identities = 44/116 (37%), Positives = 73/116 (62%), Gaps = 1/116 (0%)
Query 1 LVISGSDDKTVKIWRG-SNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWS 59
+VISGSDDKTV++W+G N +S F L+ + + ++LHP N+YF A A+D W
Sbjct 263 VVISGSDDKTVRVWKGPENADASDFKCSHVLKSSKAPVVSMSLHPSNEYFLAGASDGLWH 322
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAALQ 115
+ ++ G+ + + +D+ S S + FHPDG++ G G DG ++IWD++ Q + L+
Sbjct 323 LVDLESGQIIKICRDIPSPCSKVQFHPDGLLAAGSGTDGAVHIWDIRTQSLASTLK 378
> tpv:TP03_0215 hypothetical protein; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=496
Score = 88.6 bits (218), Expect = 4e-18, Method: Composition-based stats.
Identities = 39/114 (34%), Positives = 65/114 (57%), Gaps = 4/114 (3%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
+V+SGSDD TV++WR DF + F L+ R + L+LHP +Y + A+D W
Sbjct 265 IVLSGSDDTTVRVWR---DFETEFKCTYVLKHHRSPVKTLSLHPSGEYVLSLASDGVWGL 321
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
I G+ + +++DL + +SL HPDG++ G +G + +WD++ Q + L
Sbjct 322 CDIDSGKVIKMHRDL-PKCNSLKIHPDGLVCIGAATNGTLQVWDIRDAQPKEPL 374
> ath:AT2G33340 nucleotide binding / ubiquitin-protein ligase;
K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=485
Score = 87.0 bits (214), Expect = 1e-17, Method: Composition-based stats.
Identities = 43/114 (37%), Positives = 62/114 (54%), Gaps = 4/114 (3%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV++ S DKTV+IWR D ++ G L ++ + +HP N YF +A+ D TW F
Sbjct 278 LVLTASADKTVRIWRNPGD--GNYACGYTLNDHSAEVRAVTVHPTNKYFVSASLDGTWCF 335
Query 61 IGIKEGRYLSVYKDLSSQ--YSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRA 112
+ G L+ D S Y++ AFHPDG+I+G G + IWD+K Q A
Sbjct 336 YDLSSGSCLAQVSDDSKNVDYTAAAFHPDGLILGTGTSQSVVKIWDVKSQANVA 389
> ath:AT1G04510 transducin family protein / WD-40 repeat family
protein; K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=523
Score = 81.6 bits (200), Expect = 5e-16, Method: Composition-based stats.
Identities = 43/115 (37%), Positives = 63/115 (54%), Gaps = 5/115 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV++ S DKTV+IW S D ++T L+ ++ + +H N YF +A+ DSTW F
Sbjct 278 LVLTASSDKTVRIWGCSED--GNYTSRHTLKDHSAEVRAVTVHATNKYFVSASLDSTWCF 335
Query 61 IGIKEGRYLSVYKDLSSQ---YSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRA 112
+ G L+ D S Y++ AFHPDG+I+G G + IWD+K Q A
Sbjct 336 YDLSSGLCLAQVTDASENDVNYTAAAFHPDGLILGTGTAQSIVKIWDVKSQANVA 390
> pfa:PFC0365w PRP19-like protein, putative; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=532
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/147 (27%), Positives = 70/147 (47%), Gaps = 33/147 (22%)
Query 1 LVISGSDDKTVKIWRG------------------------------SNDF---SSSFTLG 27
+ I+ S+DKT++IW+G +N++ + F
Sbjct 269 ICITASNDKTIRIWKGDDNNVHHNDNGDSDSDNNNNNNDNDGSHLNNNEYIYDNYQFVSA 328
Query 28 VALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLAFHPD 87
+ + + + LALHPL +YF +++ DS W ++ + + KD S + LA HPD
Sbjct 329 HVITKHKDHVTSLALHPLENYFISSSKDSMWILHDLETAKTIKTSKDNPSSFKHLAIHPD 388
Query 88 GMIMGGGGVDGNIYIWDMKGQQQRAAL 114
GM+ G D NI+I+D+K Q+ +A L
Sbjct 389 GMMFGIAAQDSNIHIYDIKSQEYKATL 415
> xla:380586 prpf19, MGC52755, nmp200; PRP19/PSO4 pre-mRNA processing
factor 19 homolog; K10599 pre-mRNA-processing factor
19 [EC:6.3.2.19]
Length=504
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 41/114 (35%), Positives = 57/114 (50%), Gaps = 7/114 (6%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV S S D T+++W + + LR G + GL+LH DY + + D W+F
Sbjct 278 LVFSSSPDSTIRVWSVPDA-----SCAQVLRAHEGAVTGLSLHATGDYLLSCSDDQYWAF 332
Query 61 IGIKEGRYLSVYKDLSS--QYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRA 112
+ GR L+ D SS + FHPDG+I G G VD I IWD+K + A
Sbjct 333 SDVHVGRVLTKVTDESSGCALTCAQFHPDGLIFGTGTVDSQIKIWDLKERSNVA 386
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 27/102 (26%), Positives = 41/102 (40%), Gaps = 10/102 (9%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST---WSF 60
+G+ D +KIW + + G G + +A Y A AA DS+ W
Sbjct 367 TGTVDSQIKIWDLKERSNVANFPG-----HSGPVSCIAFSENGYYLATAADDSSVKLWDL 421
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYI 102
+K + L + + Q SL F G + GG D +YI
Sbjct 422 RKLKNFKTLQLEE--GYQVCSLVFDQSGTYLAVGGTDIQVYI 461
> mmu:28000 Prpf19, AA617263, AL024362, D19Wsu55e, NMP200, PSO4,
Prp19, Snev; PRP19/PSO4 pre-mRNA processing factor 19 homolog
(S. cerevisiae); K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=504
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 39/108 (36%), Positives = 55/108 (50%), Gaps = 7/108 (6%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV S S D T++IW N +R + GL+LH DY +++ D W+F
Sbjct 278 LVFSASPDATIRIWSVPNTSCVQ-----VVRAHESAVTGLSLHATGDYLLSSSDDQYWAF 332
Query 61 IGIKEGRYLSVYKDLSS--QYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
I+ GR L+ D +S + FHPDG+I G G +D I IWD+K
Sbjct 333 SDIQTGRVLTKVTDETSGCSLTCAQFHPDGLIFGTGTMDSQIKIWDLK 380
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 41/102 (40%), Gaps = 10/102 (9%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST---WSF 60
+G+ D +KIW D + G I +A Y A AA DS+ W
Sbjct 367 TGTMDSQIKIW----DLKERTNVA-NFPGHSGPITSIAFSENGYYLATAADDSSVKLWDL 421
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYI 102
+K + L + D + + SL F G + GG D IYI
Sbjct 422 RKLKNFKTLQL--DNNFEVKSLIFDQSGTYLALGGTDVQIYI 461
> hsa:27339 PRPF19, NMP200, PRP19, PSO4, SNEV, UBOX4, hPSO4; PRP19/PSO4
pre-mRNA processing factor 19 homolog (S. cerevisiae);
K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=504
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 40/108 (37%), Positives = 58/108 (53%), Gaps = 7/108 (6%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV S S D T++IW N +S + +R + GL+LH DY +++ D W+F
Sbjct 278 LVFSASPDATIRIWSVPN--ASCVQV---VRAHESAVTGLSLHATGDYLLSSSDDQYWAF 332
Query 61 IGIKEGRYLSVYKDLSS--QYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
I+ GR L+ D +S + FHPDG+I G G +D I IWD+K
Sbjct 333 SDIQTGRVLTKVTDETSGCSLTCAQFHPDGLIFGTGTMDSQIKIWDLK 380
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 41/102 (40%), Gaps = 10/102 (9%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST---WSF 60
+G+ D +KIW D + G I +A Y A AA DS+ W
Sbjct 367 TGTMDSQIKIW----DLKERTNVA-NFPGHSGPITSIAFSENGYYLATAADDSSVKLWDL 421
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYI 102
+K + L + D + + SL F G + GG D IYI
Sbjct 422 RKLKNFKTLQL--DNNFEVKSLIFDQSGTYLALGGTDVQIYI 461
> dre:321544 prp19, NMP200, fb18f09, wu:fb18f09, zgc:56158; PRP19/PSO4
homolog (S. cerevisiae); K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=505
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 55/110 (50%), Gaps = 11/110 (10%)
Query 1 LVISGSDDKTVKIWR--GSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTW 58
+V S S D T+++W G N +R + GL+LH DY +++ D W
Sbjct 279 VVFSASSDSTIRVWSVTGGNCVQ-------VVRAHESAVTGLSLHATGDYLLSSSEDQYW 331
Query 59 SFIGIKEGRYLSVYKDLSS--QYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+F I+ GR L+ D ++ + FHPDG+I G G D I IWD+K
Sbjct 332 AFSDIQTGRVLTKVTDETAGCALTCAQFHPDGLIFGTGTGDSQIKIWDLK 381
> cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=492
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 40/115 (34%), Positives = 61/115 (53%), Gaps = 8/115 (6%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
IS S D +++W + D SS + V + + ++L+ DY +A+ DS W+F
Sbjct 267 ISASADSHIRVWSAT-DSSSKAIIDV----HQAPVTDISLNASGDYILSASDDSYWAFSD 321
Query 63 IKEGRYL-SVYKDLSSQYS--SLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
I+ G+ L V + SQ + S+ FHPDG+I G G D + IWD+K Q AA
Sbjct 322 IRSGKSLCKVSVEPGSQIAVHSIEFHPDGLIFGTGAADAVVKIWDLKNQTVAAAF 376
> xla:734892 hypothetical protein MGC130867; K03130 transcription
initiation factor TFIID subunit 5
Length=588
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 32/106 (30%), Positives = 50/106 (47%), Gaps = 11/106 (10%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+GS DKTV++W S G RG + LA P Y A+A D +
Sbjct 442 TGSTDKTVRLWSTQQGNSVRLFTG-----HRGPVLTLAFSPNGKYLASAGEDQRLRLWDL 496
Query 64 KEGRYLSVYKDL---SSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
G ++YK+L + SSL F PD ++ G +D ++ +WD++
Sbjct 497 ASG---TLYKELRGHTDNISSLTFSPDSNLIASGSMDNSVRVWDIR 539
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 25/102 (24%), Positives = 46/102 (45%), Gaps = 5/102 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
S S D+T ++W F +F L + D+ + HP ++Y A + D T
Sbjct 400 SASHDRTGRLWC----FDRTFPLRI-YAGHLSDVDCIKFHPNSNYLATGSTDKTVRLWST 454
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++G + ++ +LAF P+G + G D + +WD+
Sbjct 455 QQGNSVRLFTGHRGPVLTLAFSPNGKYLASAGEDQRLRLWDL 496
> mmu:102162 Taf5l, 1110005N04Rik, AI849020; TAF5-like RNA polymerase
II, p300/CBP-associated factor (PCAF)-associated factor;
K03130 transcription initiation factor TFIID subunit 5
Length=589
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 45/103 (43%), Gaps = 5/103 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+GS DKTV++W S G RG + L+ P Y A+A D +
Sbjct 443 TGSTDKTVRLWSAQQGNSVRLFTG-----HRGPVLSLSFSPNGKYLASAGEDQRLKLWDL 497
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
G + + +SLAF PD ++ +D ++ +WD++
Sbjct 498 ASGTLFKELRGHTDSITSLAFSPDSGLIASASMDNSVRVWDIR 540
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 28/105 (26%), Positives = 47/105 (44%), Gaps = 11/105 (10%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRG---DICGLALHPLNDYFAAAAADSTWSF 60
SGS D+T ++W SF LR G D+ + HP ++Y A + D T
Sbjct 401 SGSHDRTARLW--------SFDRTYPLRIYAGHLADVDCVKFHPNSNYLATGSTDKTVRL 452
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++G + ++ SL+F P+G + G D + +WD+
Sbjct 453 WSAQQGNSVRLFTGHRGPVLSLSFSPNGKYLASAGEDQRLKLWDL 497
Score = 35.8 bits (81), Expect = 0.033, Method: Composition-based stats.
Identities = 25/108 (23%), Positives = 46/108 (42%), Gaps = 11/108 (10%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST---W 58
++S S+D +++ W SFT V + + + + P + YFA+ + D T W
Sbjct 357 LLSCSEDMSIRYWD-----LGSFTNTVLYQGHAYPVWDVDISPFSLYFASGSHDRTARLW 411
Query 59 SFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
SF L +Y + + FHP+ + G D + +W +
Sbjct 412 SF---DRTYPLRIYAGHLADVDCVKFHPNSNYLATGSTDKTVRLWSAQ 456
> xla:446524 taf5l, MGC80243; TAF5-like RNA polymerase II, p300/CBP-associated
factor (PCAF)-associated factor, 65kDa; K03130
transcription initiation factor TFIID subunit 5
Length=587
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+GS DKTV++W S G RG + LA P Y A+A D +
Sbjct 441 TGSSDKTVRLWSTQQGNSVRLFTG-----HRGPVLTLAFSPNGKYLASAGEDQRLKLWDL 495
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
G + + SSL F PD ++ +D ++ +WD++
Sbjct 496 ASGTQYKELRGHTDNISSLTFSPDSSLIASASMDNSVRVWDIR 538
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/112 (24%), Positives = 50/112 (44%), Gaps = 5/112 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
S S D+T ++W F +F L + D+ + HP ++Y A ++D T
Sbjct 399 SASHDRTGRLWC----FDRTFPLRI-YAGHLSDVDCIKFHPNSNYLATGSSDKTVRLWST 453
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAALQ 115
++G + ++ +LAF P+G + G D + +WD+ Q L+
Sbjct 454 QQGNSVRLFTGHRGPVLTLAFSPNGKYLASAGEDQRLKLWDLASGTQYKELR 505
> ath:AT4G15900 PRL1; PRL1 (PLEIOTROPIC REGULATORY LOCUS 1); basal
transcription repressor/ nucleotide binding / protein binding;
K12862 pleiotropic regulator 1
Length=486
Score = 51.6 bits (122), Expect = 5e-07, Method: Composition-based stats.
Identities = 38/121 (31%), Positives = 62/121 (51%), Gaps = 16/121 (13%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V++GS D T+K W D T+ L + + + LHP + FA+A+AD+T F
Sbjct 317 VVTGSHDTTIKFW----DLRYGKTMST-LTHHKKSVRAMTLHPKENAFASASADNTKKF- 370
Query 62 GIKEGRYLSVYKDLSSQYS---SLAFHPDGMIMGGGGVDGNIYIWDMKG----QQQRAAL 114
+ +G + + LS Q + ++A + DG +M GG +G+I+ WD K QQ +
Sbjct 371 SLPKGEF--CHNMLSQQKTIINAMAVNEDG-VMVTGGDNGSIWFWDWKSGHSFQQSETIV 427
Query 115 Q 115
Q
Sbjct 428 Q 428
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 30/111 (27%), Positives = 51/111 (45%), Gaps = 5/111 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+GS D+T+KIW D ++ L + L + GLA+ + Y +A D +
Sbjct 193 TGSADRTIKIW----DVATG-VLKLTLTGHIEQVRGLAVSNRHTYMFSAGDDKQVKCWDL 247
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
++ + + Y S LA HP ++ GG D +WD++ + Q AL
Sbjct 248 EQNKVIRSYHGHLSGVYCLALHPTLDVLLTGGRDSVCRVWDIRTKMQIFAL 298
> cel:C36B1.5 prp-4; yeast PRP (splicing factor) related family
member (prp-4); K12662 U4/U6 small nuclear ribonucleoprotein
PRP4
Length=496
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 33/105 (31%), Positives = 48/105 (45%), Gaps = 5/105 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V+S S D TV +W S +G L + + +A HP + A A DSTW
Sbjct 269 VVSCSYDGTVLLW----SLSQESPIG-ELEQHPQRVSKVAFHPNGHHLATACFDSTWRMY 323
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ + L + S + +AFHPDG + GG D +WDM+
Sbjct 324 DLTTKKELLYQEGHSKSVADVAFHPDGSVALTGGHDCYGRVWDMR 368
> ath:AT1G61210 WD-40 repeat family protein / katanin p80 subunit,
putative
Length=1180
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 26/105 (24%), Positives = 51/105 (48%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV++G+ +K+W D + + A R + + HP ++ A+ ++D+
Sbjct 72 LVLAGASSGVIKLW----DVEEA-KMVRAFTGHRSNCSAVEFHPFGEFLASGSSDANLKI 126
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I++ + YK S S++ F PDG + GG+D + +WD+
Sbjct 127 WDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDL 171
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 30/104 (28%), Positives = 46/104 (44%), Gaps = 6/104 (5%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V+SG D VK+W D ++ L + G I L HPL A +AD T F
Sbjct 157 VVSGGLDNVVKVW----DLTAGKLLH-EFKFHEGPIRSLDFHPLEFLLATGSADRTVKFW 211
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
++ + + ++ S+ FHPDG + G D +Y W+
Sbjct 212 DLETFELIGSTRPEATGVRSIKFHPDGRTLFCGLDDSLKVYSWE 255
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 25/106 (23%), Positives = 43/106 (40%), Gaps = 5/106 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L I+G DD V +W S ++L + +A A A+
Sbjct 30 LFITGGDDYKVNLWAIGKPTSL-----MSLCGHTSAVDSVAFDSAEVLVLAGASSGVIKL 84
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++E + + + S S++ FHP G + G D N+ IWD++
Sbjct 85 WDVEEAKMVRAFTGHRSNCSAVEFHPFGEFLASGSSDANLKIWDIR 130
Score = 35.8 bits (81), Expect = 0.031, Method: Composition-based stats.
Identities = 25/103 (24%), Positives = 39/103 (37%), Gaps = 5/103 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
SGS D +KIW D + RG I + P + + D+ +
Sbjct 117 SGSSDANLKIW----DIRKKGCIQTYKGHSRG-ISTIRFTPDGRWVVSGGLDNVVKVWDL 171
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
G+ L +K SL FHP ++ G D + WD++
Sbjct 172 TAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSADRTVKFWDLE 214
> hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1
centriolar protein homolog A (Chlamydomonas)
Length=359
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 38/140 (27%), Positives = 55/140 (39%), Gaps = 37/140 (26%)
Query 1 LVISGSDDKTVKIWRGSN-----------------DFSSSFTLGVA-----------LRR 32
L++S SDDKTVK+W S+ DF S T A +R
Sbjct 159 LIVSASDDKTVKLWDKSSRECVHSYCEHGGFVTYVDFHPSGTCIAAAGMDNTVKVWDVRT 218
Query 33 QR---------GDICGLALHPLNDYFAAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLA 83
R + GL+ HP +Y A++DST + + EGR L +++A
Sbjct 219 HRLLQHYQLHSAAVNGLSFHPSGNYLITASSDSTLKILDLMEGRLLYTLHGHQGPATTVA 278
Query 84 FHPDGMIMGGGGVDGNIYIW 103
F G GG D + +W
Sbjct 279 FSRTGEYFASGGSDEQVMVW 298
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 25/104 (24%), Positives = 44/104 (42%), Gaps = 5/104 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
++ SDDKTVK+W ++ F+L + R C P +A+ D T
Sbjct 119 VTASDDKTVKVW-ATHRQKFLFSLSQHINWVR---CA-KFSPDGRLIVSASDDKTVKLWD 173
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ Y + + + FHP G + G+D + +WD++
Sbjct 174 KSSRECVHSYCEHGGFVTYVDFHPSGTCIAAAGMDNTVKVWDVR 217
Score = 31.2 bits (69), Expect = 0.78, Method: Composition-based stats.
Identities = 24/109 (22%), Positives = 40/109 (36%), Gaps = 5/109 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ SGS DKTV+IW + S+ R + + F A+ D T
Sbjct 75 LLASGSRDKTVRIWVPNVKGEST-----VFRAHTATVRSVHFCSDGQSFVTASDDKTVKV 129
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
++L + F PDG ++ D + +WD ++
Sbjct 130 WATHRQKFLFSLSQHINWVRCAKFSPDGRLIVSASDDKTVKLWDKSSRE 178
> ath:AT1G11160 nucleotide binding
Length=1021
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 25/105 (23%), Positives = 50/105 (47%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV++G+ +K+W D S + A R + + HP ++ A+ ++D+
Sbjct 72 LVLAGASSGVIKLW----DLEES-KMVRAFTGHRSNCSAVEFHPFGEFLASGSSDTNLRV 126
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++ + YK + S++ F PDG + GG+D + +WD+
Sbjct 127 WDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDL 171
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 48/104 (46%), Gaps = 6/104 (5%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V+SG D VK+W D ++ L + G I L HPL A +AD T F
Sbjct 157 VVSGGLDNVVKVW----DLTAGKLLH-EFKCHEGPIRSLDFHPLEFLLATGSADRTVKFW 211
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
++ + + ++ ++AFHPDG + G DG +Y W+
Sbjct 212 DLETFELIGTTRPEATGVRAIAFHPDGQTLFCGLDDGLKVYSWE 255
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 23/106 (21%), Positives = 44/106 (41%), Gaps = 5/106 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+++G DD V +W S ++L + +A + A A+
Sbjct 30 LLLTGGDDYKVNLWSIGKTTSP-----MSLCGHTSPVDSVAFNSEEVLVLAGASSGVIKL 84
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++E + + + S S++ FHP G + G D N+ +WD +
Sbjct 85 WDLEESKMVRAFTGHRSNCSAVEFHPFGEFLASGSSDTNLRVWDTR 130
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 23/103 (22%), Positives = 39/103 (37%), Gaps = 5/103 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
SGS D +++W D + RG I + P + + D+ +
Sbjct 117 SGSSDTNLRVW----DTRKKGCIQTYKGHTRG-ISTIEFSPDGRWVVSGGLDNVVKVWDL 171
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
G+ L +K SL FHP ++ G D + WD++
Sbjct 172 TAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWDLE 214
> mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar protein
homolog B (Chlamydomonas)
Length=476
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 34/140 (24%), Positives = 59/140 (42%), Gaps = 37/140 (26%)
Query 1 LVISGSDDKTVKIWRGSN-----------------DFSSSFT-----------------L 26
L++S S+DKT+KIW +N DF+ + T +
Sbjct 158 LIVSCSEDKTIKIWDTTNKQCVNNFSDSVGFANFVDFNPNGTCIASAGSDHAVKIWDIRM 217
Query 27 GVALRRQRGDICG---LALHPLNDYFAAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLA 83
L+ + CG L+ HPL + A++D T + + EGR + + + +++
Sbjct 218 NKLLQHYQVHSCGVNCLSFHPLGNSLVTASSDGTVKMLDLIEGRLIYTLQGHTGPVFTVS 277
Query 84 FHPDGMIMGGGGVDGNIYIW 103
F DG ++ GG D + IW
Sbjct 278 FSKDGELLTSGGADAQVLIW 297
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 22/106 (20%), Positives = 46/106 (43%), Gaps = 5/106 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+++ S+DK++K+W + F F +L R + P + + D T
Sbjct 116 LLVTASEDKSIKVW---SMFRQRFLY--SLYRHTHWVRCAKFSPDGRLIVSCSEDKTIKI 170
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ ++ + D + + F+P+G + G D + IWD++
Sbjct 171 WDTTNKQCVNNFSDSVGFANFVDFNPNGTCIASAGSDHAVKIWDIR 216
> xla:379889 katnb1, MGC52971; katanin p80 (WD40-containing) subunit
B 1
Length=655
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/114 (23%), Positives = 52/114 (45%), Gaps = 5/114 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+++GS +++IW D ++ L L + ++C L HP ++ A+ + D+
Sbjct 77 LIVAGSQSGSLRIW----DLEAAKILRT-LMGHKANVCSLDFHPYGEFVASGSLDTNIKL 131
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
++ + YK + L F PDG + D +I +WD+ + A L
Sbjct 132 WDVRRKGCVFRYKGHTQAVRCLRFSPDGKWLASTSDDHSIKLWDLTAGKMMAEL 185
Score = 34.3 bits (77), Expect = 0.097, Method: Composition-based stats.
Identities = 24/102 (23%), Positives = 47/102 (46%), Gaps = 6/102 (5%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
S SDD ++K+W D ++ + L +G + + HP A+ +AD T F +
Sbjct 164 STSDDHSIKLW----DLTAG-KMMAELSEHKGPVNIIEFHPNEYLLASGSADRTVRFWDL 218
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
++ + + + + ++ F DG + GG D +Y W+
Sbjct 219 EKFQLIGCTEGETIPVRAILFSNDGGCIFCGGKDALRVYGWE 260
> mmu:74187 Katnb1, 2410003J24Rik, KAT; katanin p80 (WD40-containing)
subunit B 1
Length=658
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 25/105 (23%), Positives = 48/105 (45%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+++GS ++++W D ++ L L + +IC L HP ++ A+ + D+
Sbjct 77 LIVAGSQSGSIRVW----DLEAAKILRT-LMGHKANICSLDFHPYGEFVASGSQDTNIKL 131
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I+ + Y+ S L F PDG + D + +WD+
Sbjct 132 WDIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDL 176
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 28/112 (25%), Positives = 46/112 (41%), Gaps = 11/112 (9%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGD---ICGLALHPLNDYFAAAAADST 57
V SGS D +K+W + R RG + L P + A+AA D T
Sbjct 119 FVASGSQDTNIKLW--------DIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHT 170
Query 58 WSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
+ G+ +S + + + + FHP+ ++ G D I WD++ Q
Sbjct 171 VKLWDLTAGKMMSEFPGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDLEKFQ 222
Score = 35.4 bits (80), Expect = 0.049, Method: Composition-based stats.
Identities = 25/102 (24%), Positives = 46/102 (45%), Gaps = 6/102 (5%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
S +DD TVK+W D ++ + G + + HP A+ ++D T F +
Sbjct 164 SAADDHTVKLW----DLTAGKMMS-EFPGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDL 218
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
++ + +S + S+ F+PDG + G D +Y W+
Sbjct 219 EKFQVVSCIEGEPGPVRSVLFNPDGCCLYSGCQDSLRVYGWE 260
Score = 31.6 bits (70), Expect = 0.57, Method: Composition-based stats.
Identities = 22/106 (20%), Positives = 42/106 (39%), Gaps = 5/106 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ +G DD V +W + ++L + + L+ + A + +
Sbjct 35 LLATGGDDCRVNLWSINKP-----NCIMSLTGHTSPVESVRLNTPEELIVAGSQSGSIRV 89
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++ + L + SL FHP G + G D NI +WD++
Sbjct 90 WDLEAAKILRTLMGHKANICSLDFHPYGEFVASGSQDTNIKLWDIR 135
> hsa:10300 KATNB1, KAT; katanin p80 (WD repeat containing) subunit
B 1
Length=655
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 25/105 (23%), Positives = 48/105 (45%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+++GS ++++W D ++ L L + +IC L HP ++ A+ + D+
Sbjct 77 LIVAGSQSGSIRVW----DLEAAKILRT-LMGHKANICSLDFHPYGEFVASGSQDTNIKL 131
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I+ + Y+ S L F PDG + D + +WD+
Sbjct 132 WDIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDL 176
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 28/112 (25%), Positives = 46/112 (41%), Gaps = 11/112 (9%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGD---ICGLALHPLNDYFAAAAADST 57
V SGS D +K+W + R RG + L P + A+AA D T
Sbjct 119 FVASGSQDTNIKLW--------DIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHT 170
Query 58 WSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
+ G+ +S + + + + FHP+ ++ G D I WD++ Q
Sbjct 171 VKLWDLTAGKMMSEFPGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDLEKFQ 222
Score = 35.4 bits (80), Expect = 0.049, Method: Composition-based stats.
Identities = 25/102 (24%), Positives = 46/102 (45%), Gaps = 6/102 (5%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
S +DD TVK+W D ++ + G + + HP A+ ++D T F +
Sbjct 164 SAADDHTVKLW----DLTAGKMMS-EFPGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDL 218
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
++ + +S + S+ F+PDG + G D +Y W+
Sbjct 219 EKFQVVSCIEGEPGPVRSVLFNPDGCCLYSGCQDSLRVYGWE 260
Score = 31.6 bits (70), Expect = 0.57, Method: Composition-based stats.
Identities = 22/106 (20%), Positives = 42/106 (39%), Gaps = 5/106 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ +G DD V +W + ++L + + L+ + A + +
Sbjct 35 LLATGGDDCRVNLWSINKP-----NCIMSLTGHTSPVESVRLNTPEELIVAGSQSGSIRV 89
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++ + L + SL FHP G + G D NI +WD++
Sbjct 90 WDLEAAKILRTLMGHKANICSLDFHPYGEFVASGSQDTNIKLWDIR 135
> dre:558528 wdr47, si:ch211-66k16.12; WD repeat domain 47
Length=909
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 31/110 (28%), Positives = 48/110 (43%), Gaps = 7/110 (6%)
Query 1 LVISGSDDKTVKIWRGSNDF---SSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST 57
++ SGS DKTV+ W D S +G A + + +A+ P A DS+
Sbjct 758 MIASGSQDKTVRFW----DLRVPSCVRVVGTAFQGSGSPVASVAVDPSGRLLATGQEDSS 813
Query 58 WSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKG 107
I+ GR + YK SS S+ F P + G D + + D++G
Sbjct 814 CMLYDIRGGRMVQTYKPHSSDVRSVRFSPGAHYLLTGSYDNKVIVTDLQG 863
> ath:AT5G49430 transducin family protein / WD-40 repeat family
protein
Length=1677
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 33/111 (29%), Positives = 54/111 (48%), Gaps = 8/111 (7%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
VI+GSDD+ VK+W +++ L + R GDI LA+ N + A+A+ D
Sbjct 260 VITGSDDRLVKVW----SMDTAYCLA-SCRGHEGDITDLAVSSNNIFIASASNDCVIRVW 314
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHP---DGMIMGGGGVDGNIYIWDMKGQQ 109
+ +G +SV + + +++AF P + DG IWD +G Q
Sbjct 315 RLPDGLPVSVLRGHTGAVTAIAFSPRPGSPYQLLSSSDDGTCRIWDARGAQ 365
> xla:379573 poc1a, MGC69111, pix2, wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=399
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 32/106 (30%), Positives = 51/106 (48%), Gaps = 7/106 (6%)
Query 1 LVISGSDDKTVKIW-RGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWS 59
L++S SDDKT+K+W + S + SF G + + HP AAAA D+T
Sbjct 116 LIVSASDDKTIKLWDKTSRECIQSFC------EHGGFVNFVDFHPSGTCIAAAATDNTVK 169
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I+ + + Y+ S +SL+FHP G + D + + D+
Sbjct 170 VWDIRMNKLIQHYQVHSGVVNSLSFHPSGNYLITASNDSTLKVLDL 215
Score = 32.3 bits (72), Expect = 0.33, Method: Composition-based stats.
Identities = 24/109 (22%), Positives = 42/109 (38%), Gaps = 5/109 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ S S DKTV++W S S+ A + G + ++ A+ D T
Sbjct 32 LIASASRDKTVRLWVPSVKGEST-----AFKAHTGTVRSVSFSGDGQSLVTASDDKTIKV 86
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
+ ++L + F PDG ++ D I +WD ++
Sbjct 87 WTVHRQKFLFSLNQHINWVRCAKFSPDGRLIVSASDDKTIKLWDKTSRE 135
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 22/105 (20%), Positives = 41/105 (39%), Gaps = 5/105 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
+++ SDDKT+K+W F+L + R C P +A+ D T
Sbjct 75 LVTASDDKTIKVWTVHRQ-KFLFSLNQHINWVR---CA-KFSPDGRLIVSASDDKTIKLW 129
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ + + + + FHP G + D + +WD++
Sbjct 130 DKTSRECIQSFCEHGGFVNFVDFHPSGTCIAAAATDNTVKVWDIR 174
> mmu:66722 Spag16, 4921511D23Rik, 4930524F24Rik, 4930585K05Rik,
AV261009, Pf20, Wdr29; sperm associated antigen 16
Length=310
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 21/92 (22%), Positives = 36/92 (39%), Gaps = 0/92 (0%)
Query 24 FTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLA 83
+ L R + + +HP DY + + D W +G+ +G L + S
Sbjct 21 YKLNNIFRLHELPVSCIVMHPCRDYLISCSEDRLWKMVGLPQGNVLLTGSGHTDWLSGCC 80
Query 84 FHPDGMIMGGGGVDGNIYIWDMKGQQQRAALQ 115
FHP G + D I +WD+ + L+
Sbjct 81 FHPSGSKLATSSGDSTIKLWDLNKGECTLTLE 112
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 22/111 (19%), Positives = 41/111 (36%), Gaps = 5/111 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+ S D T+K+W D + + L + H D+ A+A+ D T +
Sbjct 90 TSSGDSTIKLW----DLNKG-ECTLTLEGHNHAVWSCTWHSCGDFVASASLDMTSKIWDV 144
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
R + +S+ F P I+ D + +WD + + +L
Sbjct 145 NSERCRYTLYGHTDSVNSIEFFPFSNILLTASADKTLSVWDARTGKCEQSL 195
> mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=405
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 32/106 (30%), Positives = 50/106 (47%), Gaps = 7/106 (6%)
Query 1 LVISGSDDKTVKIW-RGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWS 59
L++S SDDKTVK+W + S + S+ G + + HP AAA D+T
Sbjct 159 LIVSASDDKTVKLWDKTSRECIHSYC------EHGGFVTYVDFHPSGTCIAAAGMDNTVK 212
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ R L Y+ S+ ++L+FHP G + D + I D+
Sbjct 213 VWDARTHRLLQHYQLHSAAVNALSFHPSGNYLITASSDSTLKILDL 258
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 26/106 (24%), Positives = 42/106 (39%), Gaps = 7/106 (6%)
Query 2 VISGSDDKTVKIWRGSND-FSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
+++ SDDKTVK+W F S T + R C P +A+ D T
Sbjct 118 LVTASDDKTVKVWSTHRQRFLFSLTQHINWVR-----CA-KFSPDGRLIVSASDDKTVKL 171
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ Y + + + FHP G + G+D + +WD +
Sbjct 172 WDKTSRECIHSYCEHGGFVTYVDFHPSGTCIAAAGMDNTVKVWDAR 217
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 24/109 (22%), Positives = 39/109 (35%), Gaps = 5/109 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ SGS DKTV+IW + S+ R + + A+ D T
Sbjct 75 LLASGSRDKTVRIWVPNVKGEST-----VFRAHTATVRSVHFCSDGQSLVTASDDKTVKV 129
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
R+L + F PDG ++ D + +WD ++
Sbjct 130 WSTHRQRFLFSLTQHINWVRCAKFSPDGRLIVSASDDKTVKLWDKTSRE 178
> ath:AT2G41500 EMB2776; nucleotide binding; K12662 U4/U6 small
nuclear ribonucleoprotein PRP4
Length=554
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 43/103 (41%), Gaps = 6/103 (5%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+ S D+T K+W+ +F G R R +A HP Y + D TW I
Sbjct 315 TASADRTAKLWKTDGTLLQTFE-GHLDRLAR-----VAFHPSGKYLGTTSYDKTWRLWDI 368
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
G L + + S +AF DG + G+D +WD++
Sbjct 369 NTGAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLR 411
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 27/104 (25%), Positives = 42/104 (40%), Gaps = 11/104 (10%)
Query 6 SDDKTVKIWRGSNDFSSSFTLGVALRRQRG---DICGLALHPLNDYFAAAAADSTWSFIG 62
S DKT ++W G L Q G + G+A A+ DS
Sbjct 358 SYDKTWRLW--------DINTGAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWD 409
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++ GR + V++ S+ F P+G + GG D IWD++
Sbjct 410 LRTGRSILVFQGHIKPVFSVNFSPNGYHLASGGEDNQCRIWDLR 453
> xla:734519 poc1b, MGC114911, pix1, tuwd12, wdr51b; POC1 centriolar
protein homolog B (Chlamydomonas)
Length=468
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 32/105 (30%), Positives = 52/105 (49%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ S SDDKTV+IW D ++ + + +G + + + A+A ADST
Sbjct 158 LIASCSDDKTVRIW----DLTNRLCINTFVD-YKGHSNYVDFNQMGTCVASAGADSTVKV 212
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I+ + L Y+ ++ SSL+FHP G + DG + I D+
Sbjct 213 WDIRMNKLLQHYQVHNAGVSSLSFHPSGNYLLTASSDGTLKILDL 257
Score = 31.2 bits (69), Expect = 0.79, Method: Composition-based stats.
Identities = 23/105 (21%), Positives = 41/105 (39%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV S S D+TV++W + S+ L+ + + F A+ D +
Sbjct 74 LVASSSKDRTVRLWAPNIKGEST-----VLKAHTAVVRCVNFSSDGQTFITASDDKSIKA 128
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ R+L ++ F PDG ++ D + IWD+
Sbjct 129 WNLHRQRFLFSLTQHTNWVRCARFSPDGRLIASCSDDKTVRIWDL 173
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 21/104 (20%), Positives = 41/104 (39%), Gaps = 5/104 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
I+ SDDK++K W N F +L + + P A+ + D T
Sbjct 118 ITASDDKSIKAW---NLHRQRFLF--SLTQHTNWVRCARFSPDGRLIASCSDDKTVRIWD 172
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ ++ + D + + F+ G + G D + +WD++
Sbjct 173 LTNRLCINTFVDYKGHSNYVDFNQMGTCVASAGADSTVKVWDIR 216
> ath:AT5G08390 hypothetical protein
Length=839
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/105 (25%), Positives = 49/105 (46%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV +G+ T+K+W D + + L R + + HP ++FA+ + D+
Sbjct 73 LVAAGAASGTIKLW----DLEEAKVVRT-LTGHRSNCVSVNFHPFGEFFASGSLDTNLKI 127
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I++ + YK + + L F PDG + GG D + +WD+
Sbjct 128 WDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDL 172
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 26/107 (24%), Positives = 41/107 (38%), Gaps = 13/107 (12%)
Query 4 SGSDDKTVKIW----RGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWS 59
SGS D +KIW +G T GV + R P + + D+
Sbjct 118 SGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLR---------FTPDGRWIVSGGEDNVVK 168
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ G+ L +K + SL FHP ++ G D + WD++
Sbjct 169 VWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDLE 215
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 50 AAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
AA AA T ++E + + S S+ FHP G G +D N+ IWD++
Sbjct 75 AAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIR 131
> dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin
p80 (WD repeat containing) subunit B 1
Length=694
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V++GS ++++W D ++ L L + I L HP+ +Y A+ + DS
Sbjct 78 VVAGSLSGSLRLW----DLEAAKILRT-LMGHKASISSLDFHPMGEYLASGSVDSNIKLW 132
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++ + YK + LAF PDG + D + +WD+
Sbjct 133 DVRRKGCVFRYKGHTQAVRCLAFSPDGKWLASASDDSTVKLWDL 176
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 25/105 (23%), Positives = 47/105 (44%), Gaps = 9/105 (8%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQ--RGDICGLALHPLNDYFAAAAADSTWSFI 61
SGS D +K+W G R + + LA P + A+A+ DST
Sbjct 122 SGSVDSNIKLWDVRRK-------GCVFRYKGHTQAVRCLAFSPDGKWLASASDDSTVKLW 174
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ G+ ++ + +S + + FHP+ ++ G D + +WD++
Sbjct 175 DLIAGKMITEFTSHTSAVNVVQFHPNEYLLASGSADRTVKLWDLE 219
Score = 35.8 bits (81), Expect = 0.035, Method: Composition-based stats.
Identities = 24/106 (22%), Positives = 43/106 (40%), Gaps = 5/106 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ +G +D V IW S ++L + + + + A + +
Sbjct 35 LLATGGEDCRVNIWAVSKP-----NCIMSLTGHTSAVGCIQFNSSEERVVAGSLSGSLRL 89
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++ + L + SSL FHP G + G VD NI +WD++
Sbjct 90 WDLEAAKILRTLMGHKASISSLDFHPMGEYLASGSVDSNIKLWDVR 135
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 24/103 (23%), Positives = 43/103 (41%), Gaps = 8/103 (7%)
Query 4 SGSDDKTVKIWRG-SNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
S SDD TVK+W + + FT + + HP A+ +AD T
Sbjct 164 SASDDSTVKLWDLIAGKMITEFT------SHTSAVNVVQFHPNEYLLASGSADRTVKLWD 217
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
+++ + + + S+ F+PDG + G + +Y W+
Sbjct 218 LEKFNMIGSSEGETGVVRSVLFNPDGSCLYSGSENTLRVYGWE 260
> ath:AT5G25150 TAF5; TAF5 (TBP-ASSOCIATED FACTOR 5); nucleotide
binding / transcription regulator; K03130 transcription initiation
factor TFIID subunit 5
Length=669
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRG---DICGLALHPLNDYFAAAAADSTWSF 60
S S D+T +IW S LR G D+ + HP +Y A ++D T
Sbjct 477 SCSHDRTARIW--------SMDRIQPLRIMAGHLSDVDCVQWHPNCNYIATGSSDKTVRL 528
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++ G + ++ S SLA PDG M G DG I +WD+
Sbjct 529 WDVQTGECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTIMMWDL 573
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/104 (24%), Positives = 45/104 (43%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
+ +GS DKTV++W +G R + LA+ P Y A+ D T
Sbjct 517 IATGSSDKTVRLWDVQTGECVRIFIG-----HRSMVLSLAMSPDGRYMASGDEDGTIMMW 571
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ R ++ +S SL++ +G ++ G D + +WD+
Sbjct 572 DLSTARCITPLMGHNSCVWSLSYSGEGSLLASGSADCTVKLWDV 615
Score = 34.7 bits (78), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 25/109 (22%), Positives = 48/109 (44%), Gaps = 11/109 (10%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST--- 57
V+S S D T+++W S +++ V + + P YFA+ + D T
Sbjct 432 FVLSSSADTTIRLW--STKLNANL---VCYKGHNYPVWDAQFSPFGHYFASCSHDRTARI 486
Query 58 WSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
WS I+ R ++ + S + +HP+ + G D + +WD++
Sbjct 487 WSMDRIQPLRIMAGHL---SDVDCVQWHPNCNYIATGSSDKTVRLWDVQ 532
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 43/125 (34%), Gaps = 21/125 (16%)
Query 1 LVISGSDDKTVKIW-------------RGSNDFSSSFTLGVALRR-------QRGDICGL 40
LV G D ++K+W + ND SS ++G RR G +
Sbjct 366 LVAGGFSDSSIKVWDMAKIGQAGSGALQAEND-SSDQSIGPNGRRSYTLLLGHSGPVYSA 424
Query 41 ALHPLNDYFAAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNI 100
P D+ +++AD+T K L YK + F P G D
Sbjct 425 TFSPPGDFVLSSSADTTIRLWSTKLNANLVCYKGHNYPVWDAQFSPFGHYFASCSHDRTA 484
Query 101 YIWDM 105
IW M
Sbjct 485 RIWSM 489
> ath:AT5G23430 transducin family protein / WD-40 repeat family
protein
Length=836
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/105 (25%), Positives = 49/105 (46%), Gaps = 5/105 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV +G+ T+K+W D + + L R + + HP ++FA+ + D+
Sbjct 73 LVAAGAASGTIKLW----DLEEA-KIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKI 127
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
I++ + YK + + L F PDG + GG D + +WD+
Sbjct 128 WDIRKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDL 172
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 27/107 (25%), Positives = 42/107 (39%), Gaps = 13/107 (12%)
Query 4 SGSDDKTVKIW----RGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWS 59
SGS D +KIW +G T GV + R P + + D+
Sbjct 118 SGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLR---------FTPDGRWVVSGGEDNIVK 168
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ G+ L+ +K Q SL FHP ++ G D + WD++
Sbjct 169 VWDLTAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWDLE 215
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 50 AAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
AA AA T ++E + + S S+ FHP G G +D N+ IWD++
Sbjct 75 AAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKIWDIR 131
> sce:YBR198C TAF5, TAF90; Taf5p; K03130 transcription initiation
factor TFIID subunit 5
Length=798
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 50/104 (48%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
++SGS+DKTV++W S D ++ V+ + + ++ PL YFA A+ D T
Sbjct 540 LLSGSEDKTVRLW--SMDTHTAL---VSYKGHNHPVWDVSFSPLGHYFATASHDQTARLW 594
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
L ++ + ++FHP+G + G D +WD+
Sbjct 595 SCDHIYPLRIFAGHLNDVDCVSFHPNGCYVFTGSSDKTCRMWDV 638
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 26/106 (24%), Positives = 47/106 (44%), Gaps = 6/106 (5%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V +GS DKT ++W S S LG + +A+ P + + + D +
Sbjct 624 VFTGSSDKTCRMWDVSTGDSVRLFLG-----HTAPVISIAVCPDGRWLSTGSEDGIINVW 678
Query 62 GIKEGRYLSVYKDL-SSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
I G+ L + + SL++ +G ++ GG D + +WD+K
Sbjct 679 DIGTGKRLKQMRGHGKNAIYSLSYSKEGNVLISGGADHTVRVWDLK 724
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 23/102 (22%), Positives = 44/102 (43%), Gaps = 5/102 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+ S D+T ++W + + G D+ ++ HP Y ++D T +
Sbjct 584 TASHDQTARLWSCDHIYPLRIFAG-----HLNDVDCVSFHPNGCYVFTGSSDKTCRMWDV 638
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
G + ++ ++ S+A PDG + G DG I +WD+
Sbjct 639 STGDSVRLFLGHTAPVISIAVCPDGRWLSTGSEDGIINVWDI 680
> dre:641491 taf5, MGC123129, wu:fc38g05, zgc:123129; TAF5 RNA
polymerase II, TATA box binding protein (TBP)-associated factor;
K03130 transcription initiation factor TFIID subunit 5
Length=743
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/103 (23%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
+SG D+ ++W + D + D+ HP ++Y A ++D T
Sbjct 544 VSGGHDRVARLW--ATDHYQPLRIFAG---HLADVTCTRFHPNSNYVATGSSDRTVRLWD 598
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ G + ++ SLAF P+G + G DG + +WD+
Sbjct 599 VLNGNCVRIFTGHKGPIHSLAFSPNGKFLASGSTDGRVLLWDI 641
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 20/104 (19%), Positives = 40/104 (38%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
++S S+D T+++W +FT V + + P YF + D
Sbjct 501 LLSSSEDGTIRLWS-----LQTFTCLVGYKGHNYPVWDTQFSPFGYYFVSGGHDRVARLW 555
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ L ++ + + FHP+ + G D + +WD+
Sbjct 556 ATDHYQPLRIFAGHLADVTCTRFHPNSNYVATGSSDRTVRLWDV 599
> mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase
II, TATA box binding protein (TBP)-associated factor; K03130
transcription initiation factor TFIID subunit 5
Length=801
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/103 (23%), Positives = 43/103 (41%), Gaps = 5/103 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
+SG D+ ++W + D + D+ HP ++Y A +AD T
Sbjct 602 VSGGHDRVARLW--ATDHYQPLRIFAG---HLADVNCTRFHPNSNYVATGSADRTVRLWD 656
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ G + ++ SL F P+G + G DG + +WD+
Sbjct 657 VLNGNCVRIFTGHKGPIHSLTFSPNGRFLATGATDGRVLLWDI 699
Score = 35.0 bits (79), Expect = 0.060, Method: Composition-based stats.
Identities = 21/104 (20%), Positives = 40/104 (38%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
++S S+D TV++W +FT V + + P YF + D
Sbjct 559 LLSSSEDGTVRLWS-----LQTFTCLVGYKGHNYPVWDTQFSPYGYYFVSGGHDRVARLW 613
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ L ++ + + FHP+ + G D + +WD+
Sbjct 614 ATDHYQPLRIFAGHLADVNCTRFHPNSNYVATGSADRTVRLWDV 657
> ath:AT3G16650 PP1/PP2A phosphatases pleiotropic regulator 2
(PRL2)
Length=479
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 35/117 (29%), Positives = 60/117 (51%), Gaps = 16/117 (13%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
VI+GS D T+K W D ++ + + + +ALHP + F +A+AD+ F
Sbjct 310 VITGSHDSTIKFW----DLRYGKSMA-TITNHKKTVRAMALHPKENDFVSASADNIKKF- 363
Query 62 GIKEGRY----LSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ--QRA 112
+ +G + LS+ +D+ +++A + DG +M GG G ++ WD K QRA
Sbjct 364 SLPKGEFCHNMLSLQRDI---INAVAVNEDG-VMVTGGDKGGLWFWDWKSGHNFQRA 416
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 28/107 (26%), Positives = 49/107 (45%), Gaps = 5/107 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+GS D+T+KIW D ++ L + L G + GLA+ + Y +A D +
Sbjct 187 TGSADRTIKIW----DVATG-VLKLTLTGHIGQVRGLAVSNRHTYMFSAGDDKQVKCWDL 241
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQ 110
++ + + Y LA HP ++ GG D +WD++ + Q
Sbjct 242 EQNKVIRSYHGHLHGVYCLALHPTLDVVLTGGRDSVCRVWDIRTKMQ 288
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 43/116 (37%), Gaps = 12/116 (10%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICG---LALHPLNDYFAAAAADSTW 58
+ S DDK VK W +R G + G LALHP D DS
Sbjct 227 MFSAGDDKQVKCW--------DLEQNKVIRSYHGHLHGVYCLALHPTLDVVLTGGRDSVC 278
Query 59 SFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
I+ + V S +S LA D ++ G D I WD++ + A +
Sbjct 279 RVWDIRTKMQIFVLPHDSDVFSVLARPTDPQVITGSH-DSTIKFWDLRYGKSMATI 333
> tpv:TP04_0666 hypothetical protein; K12862 pleiotropic regulator
1
Length=521
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 35/118 (29%), Positives = 55/118 (46%), Gaps = 12/118 (10%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADS--TWS 59
VISGS DKTV++W D S ++ V L + I +++HP F + A+D+ W
Sbjct 350 VISGSQDKTVRLW----DLSMGKSI-VTLTNHKKSIRAMSIHPTEYSFCSCASDNVKVWK 404
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDG--MIMGGGGVDGNIYIWDMKGQQQRAALQ 115
EG+++ +S + A DG I+ G DG ++ WD + LQ
Sbjct 405 ---CPEGQFIRNITGHNSILNCSAIKDDGDSSILVAGSNDGQLHFWDWNSGYKFQTLQ 459
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 27/107 (25%), Positives = 52/107 (48%), Gaps = 5/107 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
+SGS D+ +KIW D +S L ++L + + + + Y + + D+T
Sbjct 225 VSGSADRLIKIW----DLASC-ELKLSLTGHINTVRDIKISTRSPYIFSCSEDNTVKCWD 279
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
I++ + + Y S L+ HP+ I+ GG D + +WD++ +Q
Sbjct 280 IEQNKVVRSYHGHLSGVYKLSLHPELDILFSGGRDAVVRVWDIRTKQ 326
> hsa:6877 TAF5, TAF2D, TAFII100; TAF5 RNA polymerase II, TATA
box binding protein (TBP)-associated factor, 100kDa; K03130
transcription initiation factor TFIID subunit 5
Length=800
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/103 (23%), Positives = 43/103 (41%), Gaps = 5/103 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
+SG D+ ++W + D + D+ HP ++Y A +AD T
Sbjct 601 VSGGHDRVARLW--ATDHYQPLRIFAG---HLADVNCTRFHPNSNYVATGSADRTVRLWD 655
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ G + ++ SL F P+G + G DG + +WD+
Sbjct 656 VLNGNCVRIFTGHKGPIHSLTFSPNGRFLATGATDGRVLLWDI 698
Score = 34.7 bits (78), Expect = 0.070, Method: Composition-based stats.
Identities = 21/104 (20%), Positives = 40/104 (38%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
++S S+D TV++W +FT V + + P YF + D
Sbjct 558 LLSSSEDGTVRLWS-----LQTFTCLVGYKGHNYPVWDTQFSPYGYYFVSGGHDRVARLW 612
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
+ L ++ + + FHP+ + G D + +WD+
Sbjct 613 ATDHYQPLRIFAGHLADVNCTRFHPNSNYVATGSADRTVRLWDV 656
> dre:567031 WD repeat-containing protein 47-like
Length=871
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 31/110 (28%), Positives = 46/110 (41%), Gaps = 7/110 (6%)
Query 1 LVISGSDDKTVKIWRGSNDF---SSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST 57
++ SGS DKTV+ W D S +G L + +A+ P A D
Sbjct 720 MIASGSQDKTVRFW----DLRVPSCVKVVGTTLHGTGSAVASVAIDPSGRLMATGLEDCR 775
Query 58 WSFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKG 107
I+ GR + VY+ SS S+ F P + G D + I D++G
Sbjct 776 CMLYDIRGGRSVQVYRPHSSDVRSVRFSPGAHYLLTGSYDSRVMISDLQG 825
> xla:398032 katnb1; katanin p80 (WD repeat containing) subunit
B 1
Length=655
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/114 (21%), Positives = 51/114 (44%), Gaps = 5/114 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+++GS +++IW D ++ L L + ++ L HP ++ A+ + D+
Sbjct 77 LIVAGSQSGSLRIW----DLEAAKILRT-LMGHKANVSSLDFHPYGEFVASGSLDTNIKL 131
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
++ + YK + L F PDG + D ++ +WD+ + A L
Sbjct 132 WDVRRKGCVFRYKGHTQAVRCLRFSPDGKWLASASDDHSVKLWDLTAGKMMAEL 185
Score = 35.0 bits (79), Expect = 0.057, Method: Composition-based stats.
Identities = 25/102 (24%), Positives = 47/102 (46%), Gaps = 6/102 (5%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
S SDD +VK+W D ++ + L +G + + HP A+ +AD T F +
Sbjct 164 SASDDHSVKLW----DLTAG-KMMAELSEHKGPVNIIEFHPNEYLLASGSADRTVRFWDL 218
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDG-NIYIWD 104
++ + + + + ++ F DG + GG D +Y W+
Sbjct 219 EKFQLVGCTEGETIPVRAILFSNDGGCIFCGGKDSLRVYGWE 260
Score = 34.3 bits (77), Expect = 0.087, Method: Composition-based stats.
Identities = 23/111 (20%), Positives = 44/111 (39%), Gaps = 9/111 (8%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQ--RGDICGLALHPLNDYFAAAAADSTW 58
V SGS D +K+W G R + + L P + A+A+ D +
Sbjct 119 FVASGSLDTNIKLWDVRRK-------GCVFRYKGHTQAVRCLRFSPDGKWLASASDDHSV 171
Query 59 SFIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
+ G+ ++ + + + FHP+ ++ G D + WD++ Q
Sbjct 172 KLWDLTAGKMMAELSEHKGPVNIIEFHPNEYLLASGSADRTVRFWDLEKFQ 222
> hsa:282809 POC1B, FLJ14923, FLJ41111, PIX1, TUWD12, WDR51B;
POC1 centriolar protein homolog B (Chlamydomonas)
Length=436
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 31/140 (22%), Positives = 55/140 (39%), Gaps = 37/140 (26%)
Query 1 LVISGSDDKTVKIWRGSN-----------------DFSSSFT------------------ 25
L++S S+DKT+KIW +N DF+ S T
Sbjct 116 LIVSCSEDKTIKIWDTTNKQCVNNFSDSVGFANFVDFNPSGTCIASAGSDQTVKVWDVRV 175
Query 26 --LGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGIKEGRYLSVYKDLSSQYSSLA 83
L + G + ++ HP +Y A++D T + + EGR + + + +++
Sbjct 176 NKLLQHYQVHSGGVNCISFHPSGNYLITASSDGTLKILDLLEGRLIYTLQGHTGPVFTVS 235
Query 84 FHPDGMIMGGGGVDGNIYIW 103
F G + GG D + +W
Sbjct 236 FSKGGELFASGGADTQVLLW 255
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 26/110 (23%), Positives = 45/110 (40%), Gaps = 7/110 (6%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L+ S S D+TV++W D F+ + + + + A A+ D +
Sbjct 32 LLASASRDRTVRLW--IPDKRGKFS---EFKAHTAPVRSVDFSADGQFLATASEDKSIKV 86
Query 61 IGIKEGRYL-SVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQ 109
+ R+L S+Y+ + F PDG ++ D I IWD +Q
Sbjct 87 WSMYRQRFLYSLYRH-THWVRCAKFSPDGRLIVSCSEDKTIKIWDTTNKQ 135
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 19/74 (25%), Positives = 35/74 (47%), Gaps = 2/74 (2%)
Query 37 ICGLALHPLNDYFAAAAADSTWS-FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGG 95
+ + P + A+A+ D T +I K G++ S +K ++ S+ F DG +
Sbjct 21 VTSVQFSPHGNLLASASRDRTVRLWIPDKRGKF-SEFKAHTAPVRSVDFSADGQFLATAS 79
Query 96 VDGNIYIWDMKGQQ 109
D +I +W M Q+
Sbjct 80 EDKSIKVWSMYRQR 93
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 19/103 (18%), Positives = 42/103 (40%), Gaps = 5/103 (4%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGI 63
+ S+DK++K+W + + F +L R + P + + D T
Sbjct 77 TASEDKSIKVW---SMYRQRFLY--SLYRHTHWVRCAKFSPDGRLIVSCSEDKTIKIWDT 131
Query 64 KEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+ ++ + D + + F+P G + G D + +WD++
Sbjct 132 TNKQCVNNFSDSVGFANFVDFNPSGTCIASAGSDQTVKVWDVR 174
> dre:556760 wdr69, si:dkey-223n17.5, zgc:153626, zgc:153780;
WD repeat domain 69
Length=418
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 29/113 (25%), Positives = 53/113 (46%), Gaps = 6/113 (5%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALH-PLNDYFAAAAADSTWSFI 61
I+GS D+T KIW D +S L L R + +A + P D A + D T
Sbjct 108 ITGSYDRTCKIW----DTASGEELHT-LEGHRNVVYAIAFNNPYGDKVATGSFDKTCKLW 162
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
+ G+ ++ +++ LAF+P ++ G +D +WD++ ++ + L
Sbjct 163 SAETGKCFYTFRGHTAEIVCLAFNPQSTLVATGSMDTTAKLWDVESGEEVSTL 215
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 45/104 (43%), Gaps = 5/104 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
V +GS DKT K+W S + F R +I LA +P + A + D+T
Sbjct 150 VATGSFDKTCKLW--SAETGKCF---YTFRGHTAEIVCLAFNPQSTLVATGSMDTTAKLW 204
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++ G +S ++ SL F+ G + G D +WD+
Sbjct 205 DVESGEEVSTLAGHFAEIISLCFNTTGDRLVTGSFDHTAILWDV 248
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 46/114 (40%), Gaps = 5/114 (4%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
LV +GS D T K+W D S + L +I L + D + D T
Sbjct 191 LVATGSMDTTAKLW----DVESGEEVST-LAGHFAEIISLCFNTTGDRLVTGSFDHTAIL 245
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAAL 114
+ GR + V + S + F+ D ++ +D + +WD +G Q A L
Sbjct 246 WDVPSGRKVHVLSGHRGEISCVQFNWDCSLIATASLDKSCKVWDAEGGQCLATL 299
> ath:AT4G02730 transducin family protein / WD-40 repeat family
protein
Length=333
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 29/105 (27%), Positives = 48/105 (45%), Gaps = 4/105 (3%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
S SDD T++IW D S + LR + + +P ++ + + D T
Sbjct 100 TCSASDDCTLRIW----DARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIW 155
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
+K G+ + + K S SS+ F+ DG ++ DG+ IWD K
Sbjct 156 EVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAK 200
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 48/118 (40%), Gaps = 10/118 (8%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSF 60
L++S S D + KIW D L + + + P + A DST
Sbjct 184 LIVSASHDGSCKIW----DAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKL 239
Query 61 IGIKEGRYLSVYKDLSSQYSSL--AFH-PDGMIMGGGGVDGNIYIWDMKGQQQRAALQ 115
G++L VY +++ + AF +G + G D +Y+WD+ Q R LQ
Sbjct 240 SNYATGKFLKVYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDL---QARNILQ 294
> cpv:cgd6_4850 PRP19 non-snRNP sliceosome component required
for DNA repair ; K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=576
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 30/100 (30%), Positives = 56/100 (56%), Gaps = 10/100 (10%)
Query 19 DFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADS-TWSFIGIKEGRYLSVYK-DLS 76
DF+SSF + I L HPL + A +++ +S ++ + L +++ + +
Sbjct 342 DFNSSFESNII-------INSLEKHPLGMHIIANSSNKGLFSMFDLESRKQLFLHQLEQN 394
Query 77 SQYSSLAFHPDGMIMGGGGVDGNIY-IWDMKGQQQRAALQ 115
+ YS L HPDG+I+GG +GNI IWD++ ++ ++L+
Sbjct 395 NSYSQLKPHPDGLILGGIVPNGNIVDIWDIRNLEKISSLE 434
> dre:100004402 plrg1, MGC174842, MGC55651, MGC86603, wu:fi74f01,
zgc:55651, zgc:86603; pleiotropic regulator 1; K12862 pleiotropic
regulator 1
Length=511
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 32/120 (26%), Positives = 60/120 (50%), Gaps = 15/120 (12%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADS--TWS 59
V++GS D T+++W D + T L + + L LHP FA+ + D+ W
Sbjct 342 VVTGSHDTTIRLW----DLVAGKTRA-TLTNHKKSVRALVLHPRQYTFASGSPDNIKQWK 396
Query 60 FIGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKG----QQQRAALQ 115
F +G ++ ++ ++LA + DG+++ G +G I++WD + Q+ AA+Q
Sbjct 397 F---PDGNFIQNLSGHNAIINTLAVNSDGVLVSGAD-NGTIHMWDWRTGYNFQRIHAAVQ 452
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 23/104 (22%), Positives = 44/104 (42%), Gaps = 5/104 (4%)
Query 3 ISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIG 62
++GS D+T+KIW D +S L ++L + G+A+ + Y + D
Sbjct 217 VTGSADRTIKIW----DLASG-KLKLSLTGHISTVRGVAVSNRSPYLFSCGEDKQVKCWD 271
Query 63 IKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
++ + + Y S L HP ++ D +WD++
Sbjct 272 LEYNKVIRHYHGHLSAVYDLDLHPTIDVLVTCSRDATARVWDIR 315
> dre:334055 taf5l, fi28g02, wu:fi28g02, zgc:63765; TAF5-like
RNA polymerase II, p300/CBP-associated factor (PCAF)-associated
factor; K03130 transcription initiation factor TFIID subunit
5
Length=601
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 26/105 (24%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Query 4 SGSDDKTVKIWRGSNDFSSSFTLGVALRRQRG---DICGLALHPLNDYFAAAAADSTWSF 60
+ S D+T ++W SF LR G D+ + HP ++Y A + D T
Sbjct 413 TASHDRTARLW--------SFARTYPLRLYAGHLSDVDCVKFHPNSNYIATGSTDKTVRL 464
Query 61 IGIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
++G + ++ +LAF P+G + G D + +WD+
Sbjct 465 WSTRQGASVRLFTGHRGPVLTLAFSPNGKYLASAGEDQRLKLWDL 509
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 26/105 (24%), Positives = 43/105 (40%), Gaps = 5/105 (4%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
++S S+D TV+ W SFT V R + + + P + YF+ A+ D T
Sbjct 369 LLSCSEDSTVRYWD-----LKSFTNTVLYRGHAYPVWDVDVSPCSLYFSTASHDRTARLW 423
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
L +Y S + FHP+ + G D + +W +
Sbjct 424 SFARTYPLRLYAGHLSDVDCVKFHPNSNYIATGSTDKTVRLWSTR 468
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 26/114 (22%), Positives = 45/114 (39%), Gaps = 11/114 (9%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFI 61
+ +GS DKTV++W S G RG + LA P Y A+A D
Sbjct 453 IATGSTDKTVRLWSTRQGASVRLFTG-----HRGPVLTLAFSPNGKYLASAGEDQRLKLW 507
Query 62 GIKEGRYLSVYKDLSSQYSSLAFHPDGM---IMGGGGVDGNIYIWDMKGQQQRA 112
+ G ++KDL +++ ++ +D + +WD++ A
Sbjct 508 DLASG---GLFKDLRGHTDTISSLSFSQDSSLVASASMDNTVRVWDIRNAHGSA 558
> bbo:BBOV_III009950 17.m07862; WD domain, G-beta repeat domain
containing protein; K05236 coatomer protein complex, subunit
alpha (xenin)
Length=1266
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 33/104 (31%), Positives = 52/104 (50%), Gaps = 20/104 (19%)
Query 2 VISGSDDKTVKIWR--GSNDFSSSFTLGVALRRQRGDICGLALHPLN-DYFAAAAADST- 57
VI+ SDDKT+++WR G N + ++ LR + +IC L +HP N +Y + + D T
Sbjct 242 VITASDDKTIRVWRYNGPNIWQTNI-----LRGHKDNICSLIMHPNNINYMISVSEDKTI 296
Query 58 --WS----FIGI----KEGRYLSVYKDLSSQYSSLAFHPDGMIM 91
W F+ KE R+ V + +S Y + H G I+
Sbjct 297 KVWDTRKWFLAYTYTSKENRFWIVQQSKNSNYIATG-HDSGFIV 339
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 29/109 (26%), Positives = 47/109 (43%), Gaps = 13/109 (11%)
Query 1 LVISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST--- 57
L +SG DD V +W DF L AL+ + + HP + +A+ D T
Sbjct 65 LFVSGGDDTHVVVW----DFRQKKML-FALKGHTDYVRTVQFHPNYPWILSASDDQTIRI 119
Query 58 WSFIGIKEGRY-LSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDM 105
W++ +GR +SV + + FHP ++ +D IWD+
Sbjct 120 WNW----QGRSCISVLQGHTHYVMCARFHPKEDLLVSASLDQTARIWDV 164
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 24/108 (22%), Positives = 43/108 (39%), Gaps = 11/108 (10%)
Query 11 VKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADS---TWSFIGIKEGR 67
+++W N TL G + G+ H L F + D+ W F ++ +
Sbjct 33 IQLWNYLNS-----TLVEVFEYHEGPVRGIDFHLLQPLFVSGGDDTHVVVWDF---RQKK 84
Query 68 YLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMKGQQQRAALQ 115
L K + ++ FHP+ + D I IW+ +G+ + LQ
Sbjct 85 MLFALKGHTDYVRTVQFHPNYPWILSASDDQTIRIWNWQGRSCISVLQ 132
Score = 28.1 bits (61), Expect = 8.0, Method: Composition-based stats.
Identities = 17/67 (25%), Positives = 30/67 (44%), Gaps = 8/67 (11%)
Query 2 VISGSDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADST---W 58
++S SDD+T++IW ++ + V +C HP D +A+ D T W
Sbjct 108 ILSASDDQTIRIW----NWQGRSCISVLQGHTHYVMCA-RFHPKEDLLVSASLDQTARIW 162
Query 59 SFIGIKE 65
++E
Sbjct 163 DVTVLRE 169
> pfa:PFE0540w WD-repeat protein, putative
Length=526
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 28/101 (27%), Positives = 42/101 (41%), Gaps = 5/101 (4%)
Query 6 SDDKTVKIWRGSNDFSSSFTLGVALRRQRGDICGLALHPLNDYFAAAAADSTWSFIGIKE 65
S D +K+W D + + G I L+ H D+FA+A+ D T I
Sbjct 306 SGDSKIKMW----DLVKEKCVHT-FKNSTGPIWSLSFHHQGDFFASASMDQTIRIFDINS 360
Query 66 GRYLSVYKDLSSQYSSLAFHPDGMIMGGGGVDGNIYIWDMK 106
R + + +S+ FHP + VD I IWDM+
Sbjct 361 LRQRQILRGHVDSVNSVNFHPYFRTLVSASVDKTISIWDMR 401
Lambda K H
0.320 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40