bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1463_orf1
Length=109
Score E
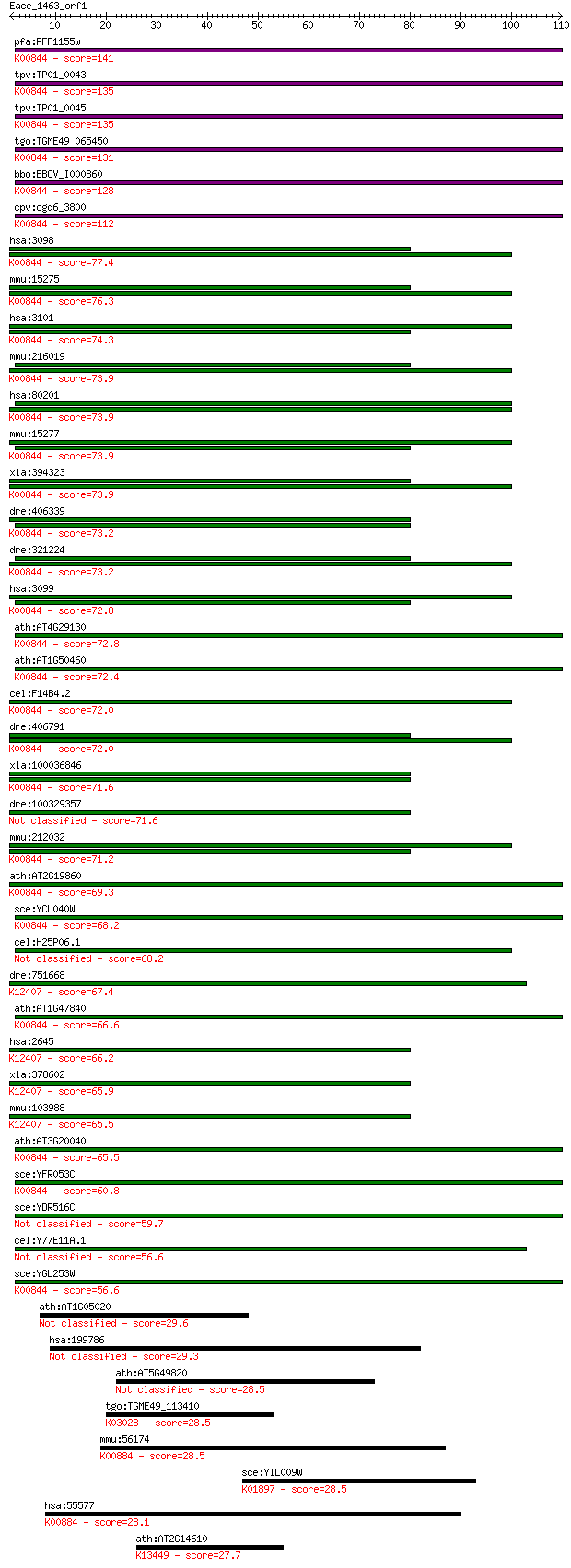
Sequences producing significant alignments: (Bits) Value
pfa:PFF1155w hexokinase (EC:2.7.1.1); K00844 hexokinase [EC:2.... 141 5e-34
tpv:TP01_0043 hexokinase; K00844 hexokinase [EC:2.7.1.1] 135 3e-32
tpv:TP01_0045 hexokinase; K00844 hexokinase [EC:2.7.1.1] 135 5e-32
tgo:TGME49_065450 hexokinase (EC:2.7.1.1); K00844 hexokinase [... 131 4e-31
bbo:BBOV_I000860 16.m00773; hexokinase (EC:2.7.1.1); K00844 he... 128 4e-30
cpv:cgd6_3800 hexokinase ; K00844 hexokinase [EC:2.7.1.1] 112 3e-25
hsa:3098 HK1, HK1-ta, HK1-tb, HK1-tc, HKI, HXK1; hexokinase 1 ... 77.4 1e-14
mmu:15275 Hk1, BB404130, Hk-1, Hk1-s, dea, mHk1-s; hexokinase ... 76.3 2e-14
hsa:3101 HK3, HKIII, HXK3; hexokinase 3 (white cell) (EC:2.7.1... 74.3 1e-13
mmu:216019 Hkdc1, BC016235, MGC28816; hexokinase domain contai... 73.9 1e-13
hsa:80201 HKDC1, FLJ22761, FLJ37767, MGC125688; hexokinase dom... 73.9 1e-13
mmu:15277 Hk2, AI642394, HKII; hexokinase 2 (EC:2.7.1.1); K008... 73.9 1e-13
xla:394323 hk1, Hexokinase, MGC80193, hk1-A; hexokinase 1 (EC:... 73.9 1e-13
dre:406339 hk2, fi09d05, fi17h06, wu:fi09d05, wu:fi17h06, wu:f... 73.2 2e-13
dre:321224 hkdc1, cb370, sb:cb370; hexokinase domain containin... 73.2 2e-13
hsa:3099 HK2, DKFZp686M1669, HKII, HXK2; hexokinase 2 (EC:2.7.... 72.8 2e-13
ath:AT4G29130 HXK1; HXK1 (HEXOKINASE 1); ATP binding / fructok... 72.8 3e-13
ath:AT1G50460 HKL1; HKL1 (HEXOKINASE-LIKE 1); ATP binding / fr... 72.4 4e-13
cel:F14B4.2 hypothetical protein; K00844 hexokinase [EC:2.7.1.1] 72.0 4e-13
dre:406791 hk1, im:7148527, wu:fc09d08, wu:fc16e02, wu:fc21e02... 72.0 5e-13
xla:100036846 hk2; hexokinase 2 (EC:2.7.1.1); K00844 hexokinas... 71.6 5e-13
dre:100329357 hexokinase 2-like 71.6 6e-13
mmu:212032 Hk3, HK_III, HK-III, MGC143940; hexokinase 3 (EC:2.... 71.2 8e-13
ath:AT2G19860 HXK2; HXK2 (HEXOKINASE 2); ATP binding / fructok... 69.3 3e-12
sce:YCL040W GLK1, HOR3; Glk1p (EC:2.7.1.2 2.7.1.1); K00844 hex... 68.2 6e-12
cel:H25P06.1 hypothetical protein 68.2 6e-12
dre:751668 gck, MGC153643, fb57f10, glk, hex4, wu:fb57f10, zgc... 67.4 1e-11
ath:AT1G47840 HXK3; HXK3 (HEXOKINASE 3); ATP binding / fructok... 66.6 2e-11
hsa:2645 GCK, FGQTL3, GK, GLK, HHF3, HK4, HKIV, HXKP, LGLK, MO... 66.2 2e-11
xla:378602 gck, gck-A, glk, hhf3, hk4, hkiv, hxkp, mody2; gluc... 65.9 3e-11
mmu:103988 Gck, GLK, Gk, HK4, HKIV, HXKP, MODY2; glucokinase (... 65.5 4e-11
ath:AT3G20040 ATHXK4; ATHXK4; ATP binding / fructokinase/ gluc... 65.5 4e-11
sce:YFR053C HXK1; Hexokinase isoenzyme 1, a cytosolic protein ... 60.8 1e-09
sce:YDR516C EMI2; Emi2p (EC:2.7.1.2) 59.7 3e-09
cel:Y77E11A.1 hypothetical protein 56.6 2e-08
sce:YGL253W HXK2, HEX1, HKB, SCI2; Hexokinase isoenzyme 2 that... 56.6 2e-08
ath:AT1G05020 epsin N-terminal homology (ENTH) domain-containi... 29.6 2.8
hsa:199786 FAM129C, BCNP1, FLJ39802; family with sequence simi... 29.3 3.0
ath:AT5G49820 emb1879 (embryo defective 1879) 28.5 4.9
tgo:TGME49_113410 26S proteasome non-ATPase regulatory subunit... 28.5 5.2
mmu:56174 Nagk, Gnk; N-acetylglucosamine kinase (EC:2.7.1.59);... 28.5 5.6
sce:YIL009W FAA3; Faa3p (EC:6.2.1.3); K01897 long-chain acyl-C... 28.5 6.1
hsa:55577 NAGK, GNK, HSA242910; N-acetylglucosamine kinase (EC... 28.1 6.7
ath:AT2G14610 PR1; PR1 (PATHOGENESIS-RELATED GENE 1); K13449 p... 27.7 9.2
> pfa:PFF1155w hexokinase (EC:2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=493
Score = 141 bits (356), Expect = 5e-34, Method: Composition-based stats.
Identities = 61/108 (56%), Positives = 79/108 (73%), Gaps = 0/108 (0%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGF TG T+DPV G+DV +L+ A R ++P + V+ND VGTL+SCAYQKGR
Sbjct 199 WTKGFETGRATNDPVEGRDVCKLMNDAFVRAAIPAKVCCVLNDAVGTLMSCAYQKGRGTP 258
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKGSIINVECGNFNKHLPTTPVD 109
C IG+ILGTG+N CY EP+ ++ Y G IIN+E GNF+K LPT+P+D
Sbjct 259 PCYIGIILGTGSNGCYYEPEWKKYKYAGKIINIEFGNFDKDLPTSPID 306
> tpv:TP01_0043 hexokinase; K00844 hexokinase [EC:2.7.1.1]
Length=506
Score = 135 bits (340), Expect = 3e-32, Method: Composition-based stats.
Identities = 62/108 (57%), Positives = 75/108 (69%), Gaps = 0/108 (0%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTK F TG T+D V GKDV L+ A KR ++ E + V+NDTVGTLLSCAYQK +
Sbjct 220 WTKDFETGRATNDQVEGKDVAILMNQAFKRNNMNAEVSIVLNDTVGTLLSCAYQKPKDYP 279
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKGSIINVECGNFNKHLPTTPVD 109
C +GVILGTG N CY E + +FGY G +IN+ECGNF+ LP TPVD
Sbjct 280 PCRVGVILGTGFNICYEEDEYERFGYVGRVINIECGNFDNELPQTPVD 327
> tpv:TP01_0045 hexokinase; K00844 hexokinase [EC:2.7.1.1]
Length=485
Score = 135 bits (339), Expect = 5e-32, Method: Composition-based stats.
Identities = 60/108 (55%), Positives = 75/108 (69%), Gaps = 0/108 (0%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTK F TG T+D V GKDV L+ A KR ++ E + V+NDTVGTLLSCAYQK +
Sbjct 199 WTKDFETGRATNDQVEGKDVAILMNQAFKRNNMNAEVSIVLNDTVGTLLSCAYQKPKDYP 258
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKGSIINVECGNFNKHLPTTPVD 109
C +GVILGTG N CY+E + +FGY G ++N+ECGNF+ LP PVD
Sbjct 259 PCRVGVILGTGFNICYVEDEYERFGYVGRVVNIECGNFDTELPLNPVD 306
> tgo:TGME49_065450 hexokinase (EC:2.7.1.1); K00844 hexokinase
[EC:2.7.1.1]
Length=468
Score = 131 bits (330), Expect = 4e-31, Method: Composition-based stats.
Identities = 57/108 (52%), Positives = 78/108 (72%), Gaps = 0/108 (0%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGF TG + D V GKDV LL A++R ++P C A+VNDTVGTL+SCAYQ+
Sbjct 185 WTKGFETGRENPDRVEGKDVAVLLADALQRHNVPAVCKAIVNDTVGTLVSCAYQRVPGTP 244
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKGSIINVECGNFNKHLPTTPVD 109
C +G+I+GTG N CY+EP+A+ +GY G+++N+E GNF+K LP +D
Sbjct 245 ECRVGLIIGTGFNACYVEPEASNYGYTGTVVNMEAGNFHKDLPRNEID 292
> bbo:BBOV_I000860 16.m00773; hexokinase (EC:2.7.1.1); K00844
hexokinase [EC:2.7.1.1]
Length=487
Score = 128 bits (322), Expect = 4e-30, Method: Composition-based stats.
Identities = 57/108 (52%), Positives = 74/108 (68%), Gaps = 0/108 (0%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGF TG DT + V G+D+ L+ A KR + + ++NDTVGTL+S AYQK
Sbjct 199 WTKGFETGRDTTEQVEGRDIGMLMDEAFKRNHVNARVSIILNDTVGTLMSVAYQKPPGYP 258
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKGSIINVECGNFNKHLPTTPVD 109
C +G+ILGTG N CY+E D +GY G ++N+ECGNF+K LPTTPVD
Sbjct 259 ECRMGLILGTGFNICYVEHDYLHYGYIGKVVNIECGNFDKLLPTTPVD 306
> cpv:cgd6_3800 hexokinase ; K00844 hexokinase [EC:2.7.1.1]
Length=518
Score = 112 bits (280), Expect = 3e-25, Method: Composition-based stats.
Identities = 57/127 (44%), Positives = 72/127 (56%), Gaps = 19/127 (14%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQ------ 55
WTK TG TDDPV GKD+ +LL A KR +P +C V+NDTVGTL+S Y
Sbjct 201 WTKEIETGRLTDDPVEGKDIGDLLNLAFKRNGVPAQCKCVLNDTVGTLISAMYDLNINNY 260
Query 56 ------------KGRSAAACTIGVILGTGANCCYIEPDAAQFGYKGSIINVECGNF-NKH 102
S IG+++GTG N CY+EP+++ FGYKG IIN ECG+F +
Sbjct 261 CDNNHSLKVNFPNNISENQPLIGIVVGTGVNACYLEPNSSNFGYKGVIINTECGDFYSTK 320
Query 103 LPTTPVD 109
LP T D
Sbjct 321 LPITDCD 327
> hsa:3098 HK1, HK1-ta, HK1-tb, HK1-tc, HKI, HXK1; hexokinase
1 (EC:2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=917
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 41/80 (51%), Positives = 54/80 (67%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTKGF TD VG DVV LL AIKR+ L+ AVVNDTVGT+++CAY++
Sbjct 618 TWTKGFKA---TD--CVGHDVVTLLRDAIKRREEFDLDVVAVVNDTVGTMMTCAYEE--- 669
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+I+GTG+N CY+E
Sbjct 670 -PTCEVGLIVGTGSNACYME 688
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 45/106 (42%), Positives = 60/106 (56%), Gaps = 18/106 (16%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTK F A+G V G DVV+LL AIK++ AVVNDTVGT+++C Y
Sbjct 170 TWTKRFKASG------VEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQH 223
Query 59 SAAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C +G+I+GTG N CY+E D + G +G + IN E G F
Sbjct 224 ----CEVGLIIGTGTNACYMEELRHIDLVE-GDEGRMCINTEWGAF 264
> mmu:15275 Hk1, BB404130, Hk-1, Hk1-s, dea, mHk1-s; hexokinase
1 (EC:2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=918
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 40/80 (50%), Positives = 54/80 (67%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTKGF TD VG DV LL A+KR+ L+ AVVNDTVGT+++CAY++
Sbjct 618 TWTKGFKA---TD--CVGHDVATLLRDAVKRREEFDLDVVAVVNDTVGTMMTCAYEE--- 669
Query 60 AAACTIGVILGTGANCCYIE 79
+C IG+I+GTG+N CY+E
Sbjct 670 -PSCEIGLIVGTGSNACYME 688
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 45/106 (42%), Positives = 61/106 (57%), Gaps = 18/106 (16%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTK F A+G V G DVV+LL AIK++ AVVNDTVGT+++C Y +
Sbjct 170 TWTKRFKASG------VEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQQ 223
Query 59 SAAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C +G+I+GTG N CY+E D + G +G + IN E G F
Sbjct 224 ----CEVGLIIGTGTNACYMEELRHIDLVE-GDEGRMCINTEWGAF 264
> hsa:3101 HK3, HKIII, HXK3; hexokinase 3 (white cell) (EC:2.7.1.1);
K00844 hexokinase [EC:2.7.1.1]
Length=923
Score = 74.3 bits (181), Expect = 1e-13, Method: Composition-based stats.
Identities = 46/104 (44%), Positives = 61/104 (58%), Gaps = 14/104 (13%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAI-KRKSLPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTKGF D + G+DVV LL AI +R+++ L A+VNDTVGT++SC Y+ R
Sbjct 624 NWTKGFKAS-DCE----GQDVVSLLREAITRRQAVELNVVAIVNDTVGTMMSCGYEDPR- 677
Query 60 AAACTIGVILGTGANCCYIEPDAAQFGYKGS----IINVECGNF 99
C IG+I+GTG N CY+E G G IN+E G F
Sbjct 678 ---CEIGLIVGTGTNACYMEELRNVAGVPGDSGRMCINMEWGAF 718
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/80 (42%), Positives = 48/80 (60%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGRS 59
SWTKGF V G+DVV+LL AI+R+ + ++ AVVNDTVGT++ C
Sbjct 183 SWTKGFRCSG-----VEGQDVVQLLRDAIRRQGAYNIDVVAVVNDTVGTMMGCE----PG 233
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+++ TG N CY+E
Sbjct 234 VRPCEVGLVVDTGTNACYME 253
> mmu:216019 Hkdc1, BC016235, MGC28816; hexokinase domain containing
1 (EC:2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=915
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 39/79 (49%), Positives = 53/79 (67%), Gaps = 10/79 (12%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF TD G+DVV++L AIKR++ L+ A+VNDTVGT+++C Y+ R
Sbjct 616 WTKGFKA---TD--CEGEDVVDMLREAIKRRNEFDLDIVAIVNDTVGTMMTCGYEDPR-- 668
Query 61 AACTIGVILGTGANCCYIE 79
C IG+I GTG+N CY+E
Sbjct 669 --CEIGLIAGTGSNVCYME 685
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 44/104 (42%), Positives = 57/104 (54%), Gaps = 14/104 (13%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSA 60
SWTK F D VV + L TA K K L ++ A+VNDTVGT+++CAY
Sbjct 170 SWTKKFKARGVQDTDVVNR----LATAMKKHKDLDVDILALVNDTVGTMMTCAYDD---- 221
Query 61 AACTIGVILGTGANCCYIEPDAAQF----GYKGSI-INVECGNF 99
C +GVI+GTG N CY+E D + G +G + IN E G F
Sbjct 222 PNCEVGVIIGTGTNACYME-DMSNIDLVEGDEGRMCINTEWGAF 264
> hsa:80201 HKDC1, FLJ22761, FLJ37767, MGC125688; hexokinase domain
containing 1 (EC:2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=917
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 45/103 (43%), Positives = 60/103 (58%), Gaps = 14/103 (13%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF TD G+DVV++L AIKR++ L+ AVVNDTVGT+++C Y+
Sbjct 618 WTKGFKA---TD--CEGEDVVDMLREAIKRRNEFDLDIVAVVNDTVGTMMTCGYED---- 668
Query 61 AACTIGVILGTGANCCYIEP----DAAQFGYKGSIINVECGNF 99
C IG+I GTG+N CY+E + + G IN E G F
Sbjct 669 PNCEIGLIAGTGSNMCYMEDMRNIEMVEGGEGKMCINTEWGGF 711
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 44/105 (41%), Positives = 59/105 (56%), Gaps = 16/105 (15%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKR-KSLPLECAAVVNDTVGTLLSCAYQKGRS 59
SWTK F D DVV LT A++R K + ++ A+VNDTVGT+++CAY
Sbjct 170 SWTKKFKARGVQD-----TDVVSRLTKAMRRHKDMDVDILALVNDTVGTMMTCAYDD--- 221
Query 60 AAACTIGVILGTGANCCYIEPDAAQF----GYKGSI-INVECGNF 99
C +GVI+GTG N CY+E D + G +G + IN E G F
Sbjct 222 -PYCEVGVIIGTGTNACYME-DMSNIDLVEGDEGRMCINTEWGAF 264
> mmu:15277 Hk2, AI642394, HKII; hexokinase 2 (EC:2.7.1.1); K00844
hexokinase [EC:2.7.1.1]
Length=917
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 45/105 (42%), Positives = 64/105 (60%), Gaps = 16/105 (15%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGRS 59
SWTKGF + V G+DVV+L+ AI+R+ ++ AVVNDTVGT+++C Y
Sbjct 170 SWTKGFKSSG-----VEGRDVVDLIRKAIQRRGDFDIDIVAVVNDTVGTMMTCGYDDQN- 223
Query 60 AAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C IG+I+GTG+N CY+E D + G +G + IN+E G F
Sbjct 224 ---CEIGLIVGTGSNACYMEEMRHIDMVE-GDEGRMCINMEWGAF 264
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 38/80 (47%), Positives = 53/80 (66%), Gaps = 12/80 (15%)
Query 2 WTKGF-ATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
WTKGF A+G + G+DVV LL AI+R+ L+ AVVNDTVGT+++C Y+
Sbjct 619 WTKGFKASGCE------GEDVVTLLKEAIRRREEFDLDVVAVVNDTVGTMMTCGYEDPH- 671
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+I+GTG+N CY+E
Sbjct 672 ---CEVGLIVGTGSNACYME 688
> xla:394323 hk1, Hexokinase, MGC80193, hk1-A; hexokinase 1 (EC:2.7.1.1);
K00844 hexokinase [EC:2.7.1.1]
Length=916
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 39/80 (48%), Positives = 53/80 (66%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTKGF + TD G+DVV LL IKR+ L+ A+VNDTVGT+++CAY+
Sbjct 618 TWTKGFKS---TD--CEGEDVVNLLREGIKRREEFDLDVVAIVNDTVGTMMTCAYED--- 669
Query 60 AAACTIGVILGTGANCCYIE 79
C IG+I+GTG+N CY+E
Sbjct 670 -PNCEIGLIVGTGSNACYME 688
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 45/106 (42%), Positives = 62/106 (58%), Gaps = 18/106 (16%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTK F A+G V G DVV+LL AIK++ + AVVNDTVGT+++C + R
Sbjct 170 TWTKRFKASG------VEGMDVVKLLNRAIKKRGDYEADIMAVVNDTVGTMMTCGFDDQR 223
Query 59 SAAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C +G+I+GTG N CY+E D + G +G + IN E G F
Sbjct 224 ----CEVGIIIGTGTNACYMEELRHIDMVE-GDEGRMCINTEWGAF 264
> dre:406339 hk2, fi09d05, fi17h06, wu:fi09d05, wu:fi17h06, wu:fi31b04,
zgc:55926; hexokinase 2 (EC:2.7.1.1); K00844 hexokinase
[EC:2.7.1.1]
Length=919
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 41/81 (50%), Positives = 51/81 (62%), Gaps = 12/81 (14%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAI-KRKSLPLECAAVVNDTVGTLLSCAYQKGR 58
SWTKGF A+G V GKDVV LL AI KR ++ AV+NDTVGT+++C Y
Sbjct 170 SWTKGFKASG------VEGKDVVSLLRKAIRKRGDFDIDIVAVINDTVGTMMTCGYDDHH 223
Query 59 SAAACTIGVILGTGANCCYIE 79
C IG+I+GTG N CY+E
Sbjct 224 ----CEIGLIVGTGTNACYME 240
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 37/80 (46%), Positives = 51/80 (63%), Gaps = 12/80 (15%)
Query 2 WTKGF-ATGHDTDDPVVGKDVVELLTAAIKR-KSLPLECAAVVNDTVGTLLSCAYQKGRS 59
WTKGF A+G + G+DV LL AI R + L+ AVVNDTVGT+++C Y+ +
Sbjct 621 WTKGFKASGCE------GEDVASLLKDAIHRCEEFDLDVVAVVNDTVGTMMTCGYEDPQ- 673
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+I+GTG N CY+E
Sbjct 674 ---CEVGLIVGTGTNACYME 690
> dre:321224 hkdc1, cb370, sb:cb370; hexokinase domain containing
1 (EC:2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=919
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 39/79 (49%), Positives = 53/79 (67%), Gaps = 10/79 (12%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF TD G DVV++L AIKR++ L+ A+VNDTVGT+++CAY+ +
Sbjct 619 WTKGFKA---TD--CEGYDVVDMLREAIKRRNEFDLDIVAIVNDTVGTMMTCAYEDPK-- 671
Query 61 AACTIGVILGTGANCCYIE 79
C IG+I GTG+N CY+E
Sbjct 672 --CQIGLIAGTGSNVCYME 688
Score = 59.7 bits (143), Expect = 3e-09, Method: Composition-based stats.
Identities = 40/105 (38%), Positives = 57/105 (54%), Gaps = 16/105 (15%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKR-KSLPLECAAVVNDTVGTLLSCAYQKGRS 59
SW+K F V G +VV+ L AI++ L ++ A+VNDTVG +++C Y
Sbjct 170 SWSKNFKV-----RGVQGTNVVQSLRKAIRKVGDLDVDVLAMVNDTVGAMMTCGYDDQN- 223
Query 60 AAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C +GVI+GTG N CY+E D + G +G + IN E G F
Sbjct 224 ---CEVGVIVGTGTNACYMEEMRHIDLVE-GDEGRMCINTELGAF 264
> hsa:3099 HK2, DKFZp686M1669, HKII, HXK2; hexokinase 2 (EC:2.7.1.1);
K00844 hexokinase [EC:2.7.1.1]
Length=917
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 45/105 (42%), Positives = 63/105 (60%), Gaps = 16/105 (15%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGRS 59
SWTKGF + V G+DVV L+ AI+R+ ++ AVVNDTVGT+++C Y
Sbjct 170 SWTKGFKSSG-----VEGRDVVALIRKAIQRRGDFDIDIVAVVNDTVGTMMTCGYDDHN- 223
Query 60 AAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C IG+I+GTG+N CY+E D + G +G + IN+E G F
Sbjct 224 ---CEIGLIVGTGSNACYMEEMRHIDMVE-GDEGRMCINMEWGAF 264
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 37/80 (46%), Positives = 53/80 (66%), Gaps = 12/80 (15%)
Query 2 WTKGF-ATGHDTDDPVVGKDVVELLTAAI-KRKSLPLECAAVVNDTVGTLLSCAYQKGRS 59
WTKGF A+G + G+DVV LL AI +R+ L+ AVVNDTVGT+++C ++
Sbjct 619 WTKGFKASGCE------GEDVVTLLKEAIHRREEFDLDVVAVVNDTVGTMMTCGFEDPH- 671
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+I+GTG+N CY+E
Sbjct 672 ---CEVGLIVGTGSNACYME 688
> ath:AT4G29130 HXK1; HXK1 (HEXOKINASE 1); ATP binding / fructokinase/
glucokinase/ hexokinase (EC:2.7.1.1); K00844 hexokinase
[EC:2.7.1.1]
Length=496
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 49/116 (42%), Positives = 61/116 (52%), Gaps = 17/116 (14%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGF+ + VG+DVV L A++R L + AA+VNDTVGTL Y A
Sbjct 193 WTKGFSI-----EEAVGQDVVGALNKALERVGLDMRIAALVNDTVGTLAGGRYYNPDVVA 247
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKG-------SIINVECGNF-NKHLPTTPVD 109
A VILGTG N Y+E A + G +IN+E GNF + HLP T D
Sbjct 248 A----VILGTGTNAAYVERATAIPKWHGLLPKSGEMVINMEWGNFRSSHLPLTEFD 299
> ath:AT1G50460 HKL1; HKL1 (HEXOKINASE-LIKE 1); ATP binding /
fructokinase/ glucokinase/ hexokinase; K00844 hexokinase [EC:2.7.1.1]
Length=498
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 44/116 (37%), Positives = 60/116 (51%), Gaps = 17/116 (14%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGF +VG+D+ E L A+ R+ L + AA+VNDTVG L Y +
Sbjct 192 WTKGFEISE-----MVGQDIAECLQGALNRRGLDMHVAALVNDTVGALSLGYYHDPDTVV 246
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYKG-------SIINVECGNF-NKHLPTTPVD 109
A V+ GTG+N CY+E A +G ++N+E GNF + HLP T D
Sbjct 247 A----VVFGTGSNACYLERTDAIIKCQGLLTTSGSMVVNMEWGNFWSSHLPRTSYD 298
> cel:F14B4.2 hypothetical protein; K00844 hexokinase [EC:2.7.1.1]
Length=495
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 50/114 (43%), Positives = 66/114 (57%), Gaps = 27/114 (23%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAI-KRKSLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTKGF A+G V G DVV LL A +RK + ++ A++NDTVGTL++CA+Q+
Sbjct 179 TWTKGFKASG------VEGVDVVTLLHEACHRRKDIDIDVVALLNDTVGTLMACAFQEN- 231
Query 59 SAAACTIGVILGTGANCCYIE-----PDAAQFGY--------KGSIINVECGNF 99
+C IGVI+GTG N CY+E P A GY + IIN E G F
Sbjct 232 ---SCQIGVIVGTGTNACYMERLDRIPKLA--GYVDEHGVTPEEMIINTEWGAF 280
> dre:406791 hk1, im:7148527, wu:fc09d08, wu:fc16e02, wu:fc21e02,
wu:fq14b11, zgc:55790, zgc:77618; hexokinase 1 (EC:2.7.1.1);
K00844 hexokinase [EC:2.7.1.1]
Length=918
Score = 72.0 bits (175), Expect = 5e-13, Method: Composition-based stats.
Identities = 38/80 (47%), Positives = 52/80 (65%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTKGF TD G+DVV LL IKR+ L+ A+VNDTVGT+++CAY++
Sbjct 618 NWTKGFKA---TD--CEGEDVVGLLREGIKRREEFDLDVVAIVNDTVGTMMTCAYEE--- 669
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+I GTG+N CY+E
Sbjct 670 -PTCEVGLIAGTGSNACYME 688
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 43/105 (40%), Positives = 60/105 (57%), Gaps = 16/105 (15%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTK F + V G DVV+LL AIK++ + AVVNDTVGT+++C + R
Sbjct 170 TWTKRFKV-----NGVEGMDVVKLLNKAIKKRGDYEADIMAVVNDTVGTMMTCGFDDQR- 223
Query 60 AAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNF 99
C +G+I+GTG N CY+E D + G +G + IN E G F
Sbjct 224 ---CEVGIIIGTGTNACYMEELRHMDMVE-GDEGRMCINTEWGAF 264
> xla:100036846 hk2; hexokinase 2 (EC:2.7.1.1); K00844 hexokinase
[EC:2.7.1.1]
Length=913
Score = 71.6 bits (174), Expect = 5e-13, Method: Composition-based stats.
Identities = 39/80 (48%), Positives = 52/80 (65%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELL-TAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRS 59
+WTKGF+ VGKDVV LL AA+++K+ + A+VNDTVGT++SC Y
Sbjct 612 TWTKGFSASG-----CVGKDVVTLLREAALRKKNNDIIVVALVNDTVGTMMSCGYND--- 663
Query 60 AAACTIGVILGTGANCCYIE 79
AC IG+I+GTG N CY+E
Sbjct 664 -PACEIGLIVGTGTNACYME 682
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 37/80 (46%), Positives = 48/80 (60%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
SWTKGF V GKDVV L AI+R+ ++ A+VNDTVG ++SC YQ
Sbjct 166 SWTKGFHCSE-----VEGKDVVHLFREAIRRQGGYDVDVIAIVNDTVGAMMSCGYQD--- 217
Query 60 AAACTIGVILGTGANCCYIE 79
+C +G I+GTG N CY+E
Sbjct 218 -HSCEVGFIVGTGTNLCYME 236
> dre:100329357 hexokinase 2-like
Length=292
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 51/81 (62%), Gaps = 12/81 (14%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAI-KRKSLPLECAAVVNDTVGTLLSCAYQKGR 58
SWTKGF A+G V GKDVV LL AI KR ++ AV+NDTVGT+++C Y
Sbjct 170 SWTKGFKASG------VEGKDVVSLLRKAIRKRGDFDIDIVAVINDTVGTMMTCGYDDHH 223
Query 59 SAAACTIGVILGTGANCCYIE 79
C IG+I+GTG N CY+E
Sbjct 224 ----CEIGLIVGTGTNACYME 240
> mmu:212032 Hk3, HK_III, HK-III, MGC143940; hexokinase 3 (EC:2.7.1.1);
K00844 hexokinase [EC:2.7.1.1]
Length=922
Score = 71.2 bits (173), Expect = 8e-13, Method: Composition-based stats.
Identities = 47/105 (44%), Positives = 65/105 (61%), Gaps = 16/105 (15%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTKGF A+G + G+DVV LL AI+R+ ++ L A+VNDTVGT++SC Y R
Sbjct 623 NWTKGFNASGCE------GQDVVYLLREAIRRRQAVELNVVAIVNDTVGTMMSCGYDDPR 676
Query 59 SAAACTIGVILGTGANCCYIEP---DAAQFGYKGSI-INVECGNF 99
C +G+I+GTG N CY+E A+ G G + IN+E G F
Sbjct 677 ----CEMGLIVGTGTNACYMEELRNVASVPGDSGLMCINMEWGAF 717
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 34/80 (42%), Positives = 48/80 (60%), Gaps = 10/80 (12%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGRS 59
SWTKGF V G+DVV+LL AI+R+ + ++ A+VNDTVGT++ C
Sbjct 181 SWTKGFRCSG-----VEGQDVVQLLRDAIQRQGTYRIDVVAMVNDTVGTMMGCEL----G 231
Query 60 AAACTIGVILGTGANCCYIE 79
C +G+I+ TG N CY+E
Sbjct 232 TRPCEVGLIVDTGTNACYME 251
> ath:AT2G19860 HXK2; HXK2 (HEXOKINASE 2); ATP binding / fructokinase/
glucokinase/ hexokinase; K00844 hexokinase [EC:2.7.1.1]
Length=393
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 47/117 (40%), Positives = 61/117 (52%), Gaps = 17/117 (14%)
Query 1 SWTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSA 60
+WTKGF+ D V KDVV L A++R L + AA+VNDT+GTL Y +
Sbjct 83 NWTKGFSI-----DDTVDKDVVGELVKAMERVGLDMLVAALVNDTIGTLAGGRY----TN 133
Query 61 AACTIGVILGTGANCCYIEPDAAQFGYKG-------SIINVECGNF-NKHLPTTPVD 109
+ VILGTG N Y+E A + G +IN+E GNF + HLP T D
Sbjct 134 PDVVVAVILGTGTNAAYVERAHAIPKWHGLLPKSGEMVINMEWGNFRSSHLPLTEYD 190
> sce:YCL040W GLK1, HOR3; Glk1p (EC:2.7.1.2 2.7.1.1); K00844 hexokinase
[EC:2.7.1.1]
Length=500
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 50/132 (37%), Positives = 64/132 (48%), Gaps = 29/132 (21%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPL-ECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF DT VGKDVV+L + + +P+ + A+ NDTVGT LS Y +
Sbjct 180 WTKGFRIA-DT----VGKDVVQLYQEQLSAQGMPMIKVVALTNDTVGTYLSHCYTSDNTD 234
Query 61 AACT-------IGVILGTGANCCYIEP-------------DAAQFGYKGSIINVECGNFN 100
+ + IG I GTG N CY+E + G IINVE G+F+
Sbjct 235 SMTSGEISEPVIGCIFGTGTNGCYMEEINKITKLPQELRDKLIKEGKTHMIINVEWGSFD 294
Query 101 ---KHLPTTPVD 109
KHLPTT D
Sbjct 295 NELKHLPTTKYD 306
> cel:H25P06.1 hypothetical protein
Length=552
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 44/110 (40%), Positives = 59/110 (53%), Gaps = 21/110 (19%)
Query 2 WTKGF-ATGHDTDDPVVGKDVVELLTAAIKRKS-LPLECAAVVNDTVGTLLSCAYQKGRS 59
WTKG ATG VG+DVV+LL AI R + + A++NDTVGT+++ AY+ G
Sbjct 193 WTKGVTATG------CVGEDVVQLLEDAIARDGRVKVNIVALINDTVGTMVAAAYEAG-- 244
Query 60 AAACTIGVILGTGANCCYIEPD----------AAQFGYKGSIINVECGNF 99
C IGVI+GTG N Y+E + +K II+ E G F
Sbjct 245 -GHCDIGVIIGTGTNASYMEDSRKIVHGLANATEDYNHKSMIIDTEWGGF 293
> dre:751668 gck, MGC153643, fb57f10, glk, hex4, wu:fb57f10, zgc:153643;
glucokinase (hexokinase 4, maturity onset diabetes
of the young 2) (EC:2.7.1.2); K12407 glucokinase [EC:2.7.1.2]
Length=476
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 43/109 (39%), Positives = 64/109 (58%), Gaps = 18/109 (16%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTKGF A+G + G +VV LL AIKR+ ++ A+VNDTV T++SC Y+
Sbjct 176 NWTKGFKASGAE------GNNVVGLLRDAIKRRGDFEMDVVAMVNDTVATMISCYYED-- 227
Query 59 SAAACTIGVILGTGANCCYIEP----DAAQFGYKGSI-INVECGNFNKH 102
+C +G+I+GTG N CY+E + + G +G + +N E G F H
Sbjct 228 --RSCEVGMIVGTGCNACYMEEMRKVELVE-GEEGRMCVNTEWGAFGDH 273
> ath:AT1G47840 HXK3; HXK3 (HEXOKINASE 3); ATP binding / fructokinase/
glucokinase/ hexokinase (EC:2.7.1.4); K00844 hexokinase
[EC:2.7.1.1]
Length=493
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 46/114 (40%), Positives = 57/114 (50%), Gaps = 15/114 (13%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGF + GK+VV L A++ L + +A+VND VGTL Y
Sbjct 197 WTKGFKVSG-----MEGKNVVACLNEAMEAHGLDMRVSALVNDGVGTLAGARYWD----E 247
Query 62 ACTIGVILGTGANCCYIEPDAA--QFGYKGS----IINVECGNFNKHLPTTPVD 109
+GVILGTG N CY+E A + K S IIN E G F+K LP T D
Sbjct 248 DVMVGVILGTGTNACYVEQKHAIPKLRSKSSSGTTIINTEWGGFSKILPQTIFD 301
> hsa:2645 GCK, FGQTL3, GK, GLK, HHF3, HK4, HKIV, HXKP, LGLK,
MODY2; glucokinase (hexokinase 4) (EC:2.7.1.2); K12407 glucokinase
[EC:2.7.1.2]
Length=465
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 36/81 (44%), Positives = 52/81 (64%), Gaps = 12/81 (14%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTKGF A+G + G +VV LL AIKR+ ++ A+VNDTV T++SC Y+ +
Sbjct 166 NWTKGFKASGAE------GNNVVGLLRDAIKRRGDFEMDVVAMVNDTVATMISCYYEDHQ 219
Query 59 SAAACTIGVILGTGANCCYIE 79
C +G+I+GTG N CY+E
Sbjct 220 ----CEVGMIVGTGCNACYME 236
> xla:378602 gck, gck-A, glk, hhf3, hk4, hkiv, hxkp, mody2; glucokinase
(hexokinase 4) (EC:2.7.1.2); K12407 glucokinase [EC:2.7.1.2]
Length=458
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 12/81 (14%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTKGF A+G + G +VV LL AIKR+ ++ A+VNDTV T++SC Y+
Sbjct 159 NWTKGFKASGAE------GNNVVGLLRDAIKRRGDFEMDVVAMVNDTVATMISCYYEDHH 212
Query 59 SAAACTIGVILGTGANCCYIE 79
C +G+I+GTG N CY+E
Sbjct 213 ----CEVGLIVGTGCNACYME 229
> mmu:103988 Gck, GLK, Gk, HK4, HKIV, HXKP, MODY2; glucokinase
(EC:2.7.1.2); K12407 glucokinase [EC:2.7.1.2]
Length=465
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 52/81 (64%), Gaps = 12/81 (14%)
Query 1 SWTKGF-ATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGR 58
+WTKGF A+G + G ++V LL AIKR+ ++ A+VNDTV T++SC Y+ +
Sbjct 166 NWTKGFKASGAE------GNNIVGLLRDAIKRRGDFEMDVVAMVNDTVATMISCYYEDRQ 219
Query 59 SAAACTIGVILGTGANCCYIE 79
C +G+I+GTG N CY+E
Sbjct 220 ----CEVGMIVGTGCNACYME 236
> ath:AT3G20040 ATHXK4; ATHXK4; ATP binding / fructokinase/ glucokinase/
hexokinase; K00844 hexokinase [EC:2.7.1.1]
Length=502
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 43/116 (37%), Positives = 61/116 (52%), Gaps = 17/116 (14%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAA 61
WTKGFA + G+D+ E L A+ ++ L + AA+VNDTVG L + + A
Sbjct 193 WTKGFAISE-----MAGEDIAECLQGALNKRGLDIRVAALVNDTVGALSFGHFHDPDTIA 247
Query 62 ACTIGVILGTGANCCYIEPDAAQFGYK------GS-IINVECGNF-NKHLPTTPVD 109
A V+ GTG+N CY+E A + GS ++N+E GNF + LP T D
Sbjct 248 A----VVFGTGSNACYLERTDAIIKCQNPRTTSGSMVVNMEWGNFWSSRLPRTSYD 299
> sce:YFR053C HXK1; Hexokinase isoenzyme 1, a cytosolic protein
that catalyzes phosphorylation of glucose during glucose metabolism;
expression is highest during growth on non-glucose
carbon sources; glucose-induced repression involves the hexokinase
Hxk2p (EC:2.7.1.4 2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=485
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 46/123 (37%), Positives = 62/123 (50%), Gaps = 25/123 (20%)
Query 2 WTKGFATGHDTDDP-VVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF D P V G DVV LL I ++ LP+E A++NDTVGTL++ Y +
Sbjct 174 WTKGF------DIPNVEGHDVVPLLQNEISKRELPIEIVALINDTVGTLIASYYTDPET- 226
Query 61 AACTIGVILGTGANCCYIEPDAAQFGYKGSI-----------INVECGNF-NKH--LPTT 106
+GVI GTG N + + + +G + IN E G+F N+H LP T
Sbjct 227 ---KMGVIFGTGVNGAFYDVVSDIEKLEGKLADDIPSNSPMAINCEYGSFDNEHLVLPRT 283
Query 107 PVD 109
D
Sbjct 284 KYD 286
> sce:YDR516C EMI2; Emi2p (EC:2.7.1.2)
Length=500
Score = 59.7 bits (143), Expect = 3e-09, Method: Composition-based stats.
Identities = 48/131 (36%), Positives = 59/131 (45%), Gaps = 28/131 (21%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRKSLPL-ECAAVVNDTVGTLLSCAYQKGRSA 60
WTK F DT VGKDVV L + + L + A+ NDTVGT LS Y G
Sbjct 181 WTKSFKI-EDT----VGKDVVRLYQEQLDIQGLSMINVVALTNDTVGTFLSHCYTSGSRP 235
Query 61 AAC------TIGVILGTGANCCYIE--------PDAAQF-----GYKGSIINVECGNFN- 100
++ IG I GTG N CY+E PD + G IN+E G+F+
Sbjct 236 SSAGEISEPVIGCIFGTGTNGCYMEDIENIKKLPDELRTRLLHEGKTQMCINIEWGSFDN 295
Query 101 --KHLPTTPVD 109
KHL T D
Sbjct 296 ELKHLSATKYD 306
> cel:Y77E11A.1 hypothetical protein
Length=451
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 42/111 (37%), Positives = 56/111 (50%), Gaps = 18/111 (16%)
Query 2 WTKGFATGHDTDDPVVGKDVVELLTAAIKRK-SLPLECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF D D + +D+V LL A++ S + AV+NDTVG L + A++ G
Sbjct 154 WTKGF----DIKD-CLQRDIVALLEEALEMNMSTKVSIKAVMNDTVGQLAAAAHKYG--- 205
Query 61 AACTIGVILGTGANCCYIEP-------DAAQFGY--KGSIINVECGNFNKH 102
CTIGV++G G N Y+E DA GY I+ E F KH
Sbjct 206 PECTIGVVIGYGCNSSYLEKTSRITKFDAKARGYDHDNMIVVTEWEEFGKH 256
> sce:YGL253W HXK2, HEX1, HKB, SCI2; Hexokinase isoenzyme 2 that
catalyzes phosphorylation of glucose in the cytosol; predominant
hexokinase during growth on glucose; functions in the
nucleus to repress expression of HXK1 and GLK1 and to induce
expression of its own gene (EC:2.7.1.4 2.7.1.1); K00844 hexokinase
[EC:2.7.1.1]
Length=486
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 42/123 (34%), Positives = 61/123 (49%), Gaps = 25/123 (20%)
Query 2 WTKGFATGHDTDDP-VVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSA 60
WTKGF D P + DVV +L I ++++P+E A++NDT GTL++ Y +
Sbjct 174 WTKGF------DIPNIENHDVVPMLQKQITKRNIPIEVVALINDTTGTLVASYYTDPET- 226
Query 61 AACTIGVILGTGANCCYIEPDAAQFGYKGSI-----------INVECGNF-NKH--LPTT 106
+GVI GTG N Y + + +G + IN E G+F N+H LP T
Sbjct 227 ---KMGVIFGTGVNGAYYDVCSDIEKLQGKLSDDIPPSAPMAINCEYGSFDNEHVVLPRT 283
Query 107 PVD 109
D
Sbjct 284 KYD 286
> ath:AT1G05020 epsin N-terminal homology (ENTH) domain-containing
protein / clathrin assembly protein-related
Length=653
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 7 ATGHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVG 47
AT HD + P+ + V E+L +KS CAA + +G
Sbjct 43 ATSHDEEVPIDDRLVTEILGIISSKKSHAASCAAAIGRRIG 83
> hsa:199786 FAM129C, BCNP1, FLJ39802; family with sequence similarity
129, member C
Length=651
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 30/73 (41%), Gaps = 0/73 (0%)
Query 9 GHDTDDPVVGKDVVELLTAAIKRKSLPLECAAVVNDTVGTLLSCAYQKGRSAAACTIGVI 68
GH DD V E+LTA + R+ LP A + G + A+ A V+
Sbjct 254 GHFGDDDVTLGSDAEVLTAVLMREQLPALRAQTLPGLRGAGRARAWAWTELLDAVHAAVL 313
Query 69 LGTGANCCYIEPD 81
G A C +P+
Sbjct 314 AGASAGLCAFQPE 326
> ath:AT5G49820 emb1879 (embryo defective 1879)
Length=497
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 29/67 (43%), Gaps = 16/67 (23%)
Query 22 VELLTAAIKRKSLPLECAAVVNDTVGTLLSC--------AYQKGRSAAACT--------I 65
VEL TAA+ LPL CAA V V + S A+ KG + T I
Sbjct 212 VELATAAVPHLFLPLACAANVVKNVAAVTSTSTRTPIYKAFAKGENIGDVTAKGECVGNI 271
Query 66 GVILGTG 72
++GTG
Sbjct 272 ADLMGTG 278
> tgo:TGME49_113410 26S proteasome non-ATPase regulatory subunit
2, putative ; K03028 26S proteasome regulatory subunit N1
Length=1091
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 20 DVVELLTAAIKRKSLPLECAAVVNDTVGTLL--SC 52
DV+E LT I SLP+EC+A ++G + SC
Sbjct 563 DVMEALTVVIVDSSLPVECSAFAAVSLGLVFVGSC 597
> mmu:56174 Nagk, Gnk; N-acetylglucosamine kinase (EC:2.7.1.59);
K00884 N-acetylglucosamine kinase [EC:2.7.1.59]
Length=292
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 32/69 (46%), Gaps = 8/69 (11%)
Query 19 KDVVELLTAAIKRKSLPL-ECAAVVNDTVGTLLSCAYQKGRSAAACTIGVILGTGANCCY 77
+D V LL ++ + L E + D G++ + G I +I GTG+NC
Sbjct 30 EDAVRLLIEELRHRFPNLSENYLITTDAAGSIATATPDGG-------IVLISGTGSNCRL 82
Query 78 IEPDAAQFG 86
I PD ++ G
Sbjct 83 INPDGSESG 91
> sce:YIL009W FAA3; Faa3p (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=694
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 7/51 (13%)
Query 47 GTLLSCAYQKGRSAAACTIGVILGTG-----ANCCYIEPDAAQFGYKGSII 92
G+ +S QK S C + I+G G AN C +EPD ++G G ++
Sbjct 430 GSAISIDAQKFFSIVLCPM--IIGYGLTETVANACVLEPDHFEYGIVGDLV 478
> hsa:55577 NAGK, GNK, HSA242910; N-acetylglucosamine kinase (EC:2.7.1.59);
K00884 N-acetylglucosamine kinase [EC:2.7.1.59]
Length=390
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 37/84 (44%), Gaps = 16/84 (19%)
Query 8 TGHDTDDPVVGKDVVELLTAAIKRKSLPL--ECAAVVNDTVGTLLSCAYQKGRSAAACTI 65
+G D +D G+ ++E L R P E + D G++ + G +
Sbjct 122 SGGDQED--AGRILIEEL-----RDRFPYLSESYLITTDAAGSIATATPDGG-------V 167
Query 66 GVILGTGANCCYIEPDAAQFGYKG 89
+I GTG+NC I PD ++ G G
Sbjct 168 VLISGTGSNCRLINPDGSESGCGG 191
> ath:AT2G14610 PR1; PR1 (PATHOGENESIS-RELATED GENE 1); K13449
pathogenesis-related protein 1
Length=161
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 26 TAAIKRKSLPLECAAVVNDTVGTLLSCAY 54
T + RKS+ L CA V + GT++SC Y
Sbjct 121 TQVVWRKSVRLGCAKVRCNNGGTIISCNY 149
Lambda K H
0.318 0.135 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40