bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1498_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
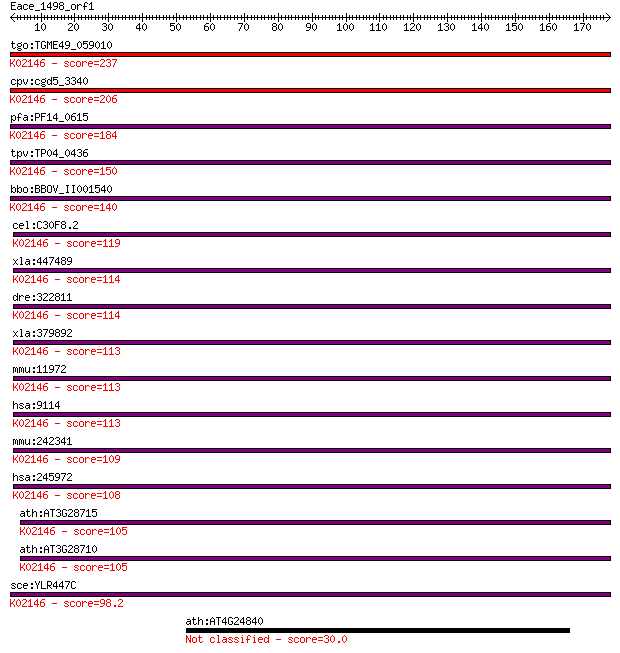
tgo:TGME49_059010 vacuolar ATP synthase subunit D, putative (E... 237 2e-62
cpv:cgd5_3340 vacuolar ATP synthase subunit d ; K02146 V-type ... 206 3e-53
pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146 ... 184 1e-46
tpv:TP04_0436 vacuolar ATP synthase (C/AC39) subunit (EC:3.6.3... 150 3e-36
bbo:BBOV_II001540 18.m06119; vacuolar ATP synthase subunit d; ... 140 2e-33
cel:C30F8.2 vha-16; Vacuolar H ATPase family member (vha-16); ... 119 7e-27
xla:447489 atp6v0d2, MGC81907; ATPase, H+ transporting, lysoso... 114 2e-25
dre:322811 atp6v0d1, fb73h07, wu:fb73h07, zgc:63769; ATPase, H... 114 2e-25
xla:379892 atp6v0d1, MGC53957, atp6d; ATPase, H+ transporting,... 113 3e-25
mmu:11972 Atp6v0d1, AI267038, Ac39, Atp6d, P39, VATX, Vma6; AT... 113 4e-25
hsa:9114 ATP6V0D1, ATP6D, ATP6DV, FLJ43534, P39, VATX, VMA6, V... 113 4e-25
mmu:242341 Atp6v0d2, 1620401A02Rik, AI324824, V-ATPase; ATPase... 109 5e-24
hsa:245972 ATP6V0D2, ATP6D2, FLJ38708, VMA6; ATPase, H+ transp... 108 9e-24
ath:AT3G28715 H+-transporting two-sector ATPase, putative; K02... 105 6e-23
ath:AT3G28710 H+-transporting two-sector ATPase, putative; K02... 105 8e-23
sce:YLR447C VMA6; Subunit d of the five-subunit V0 integral me... 98.2 1e-20
ath:AT4G24840 hypothetical protein 30.0 4.3
> tgo:TGME49_059010 vacuolar ATP synthase subunit D, putative
(EC:3.6.3.14); K02146 V-type H+-transporting ATPase subunit
AC39 [EC:3.6.3.14]
Length=396
Score = 237 bits (604), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 114/179 (63%), Positives = 140/179 (78%), Gaps = 3/179 (1%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
R K+ +EFRY++SQA PL KF+D+IA +KMIDN++GLIQGALNRK + ELLAR DP+G
Sbjct 71 CREKMASEFRYMRSQASGPLGKFMDFIATEKMIDNVVGLIQGALNRKSSHELLARVDPMG 130
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTA--AAATPDNPTGHSLRQI 118
YF EM AIA MD ++SYEELYR+LLIDTPVG+YF FL + AAA G SL ++
Sbjct 131 YFQEMAAIANMDLTTSYEELYRALLIDTPVGKYFQAFLTESGSQAAAHSAEHGGRSLAEV 190
Query 119 EHIMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
I++E D++L+RNSLKKGWLEDFY FVQ+L TTKEVMTHILK EAD+RVL L +NS
Sbjct 191 ASIVSETDIELMRNSLKKGWLEDFYAFVQSL-GGTTKEVMTHILKREADYRVLRLVVNS 248
> cpv:cgd5_3340 vacuolar ATP synthase subunit d ; K02146 V-type
H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=412
Score = 206 bits (525), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 100/177 (56%), Positives = 130/177 (73%), Gaps = 11/177 (6%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
R K EFR LQSQAY+PL KFL+YI +KMIDN++ LIQGALN+KPAEELLAR DPLG
Sbjct 88 CREKFAHEFRMLQSQAYEPLGKFLNYITYEKMIDNVVNLIQGALNKKPAEELLARLDPLG 147
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
YF E+RA +D SSS++ELY+S+LI+TP+G YFD FL + + +
Sbjct 148 YFPEIRAFVALDLSSSFDELYKSILIETPIGPYFDEFLTSFSGEN----------EDVTS 197
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
I+ E D+++LR+SLKK WLEDFY F QTL + T+ EVM+H+LK EADFR+L++ +NS
Sbjct 198 IVKEMDLEILRSSLKKSWLEDFYRFCQTL-NPTSAEVMSHVLKCEADFRLLAITLNS 253
> pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=382
Score = 184 bits (468), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 86/177 (48%), Positives = 127/177 (71%), Gaps = 10/177 (5%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
+ K+ EF Y+++QA +PL FLDYIA +KMIDN++ LIQG LN+KPA+ELL+R DPLG
Sbjct 71 CKEKMAHEFNYIRAQAEEPLRTFLDYIAKEKMIDNVISLIQGTLNKKPADELLSRVDPLG 130
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
YF +M+AI MD +SY+++ + LLI+TP+G YFD +++ ++ +
Sbjct 131 YFPQMKAITTMDVQNSYDDVLKVLLIETPIGSYFDQYISANSSNEN---------NNVTT 181
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
I+++ D+++LRN+LKK WLEDFY F++ L T+EVM HILK+ ADFRVLS+ +N+
Sbjct 182 ILSDMDIEILRNTLKKAWLEDFYNFIRKL-GGKTEEVMGHILKSVADFRVLSVTLNT 237
> tpv:TP04_0436 vacuolar ATP synthase (C/AC39) subunit (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=383
Score = 150 bits (378), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 75/177 (42%), Positives = 115/177 (64%), Gaps = 5/177 (2%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
+ KL ++++YL+ Q+ LA FLD+IA +KMIDN++ L+QG LN+ +EL+ R DP+G
Sbjct 71 CKEKLASDYQYLRQQSDGDLAVFLDFIAREKMIDNLIALLQGLLNKTDPDELMDRLDPIG 130
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
+F ++A+ + S EELYR +L DTP+G YF+ +L T + ++ +
Sbjct 131 WFRGIKALLSSEIGQSAEELYRIILCDTPIGPYFERYL----PTVTYTRGSSSNIDKTHK 186
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
I+ A++ +++ +LKK WLEDFY F +L TT +VM HILKTEADF+ LSL +N
Sbjct 187 ILDSANIAIMKATLKKMWLEDFYNFSVSL-GGTTADVMGHILKTEADFKALSLTLNC 242
> bbo:BBOV_II001540 18.m06119; vacuolar ATP synthase subunit d;
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=374
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 75/177 (42%), Positives = 106/177 (59%), Gaps = 13/177 (7%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
KL +F YL Q+ LA FLD++A +KMIDN++ L+QG NRK EEL+ R DP+G
Sbjct 71 CNEKLANDFAYLCQQSDGKLAIFLDFVAREKMIDNLIALLQGVSNRKTPEELMERVDPIG 130
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
+F + + D S E+L+R +L DTP+G YF+ L A D+ +
Sbjct 131 WFRGLETLMDADMCQSVEDLHRIILCDTPIGTYFERVL-----PAISDSKK-------QS 178
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
++ + + LL++ LKK WLEDFY F +L T+ EVM HIL+TEADFR L+L +N
Sbjct 179 VLDPSSITLLKSFLKKAWLEDFYEFSNSL-GGTSAEVMGHILRTEADFRDLALTLNC 234
> cel:C30F8.2 vha-16; Vacuolar H ATPase family member (vha-16);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=348
Score = 119 bits (297), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 69/176 (39%), Positives = 99/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ KL EF +L++ A +PLA FLDYI MIDN++ LI G L+++P EL+ + PLG
Sbjct 72 KEKLVTEFTHLRNNALEPLATFLDYITYSYMIDNIILLITGTLHQRPISELINKCHPLGS 131
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M AI +S+ ELY ++L+DTP+ YF +N E
Sbjct 132 FEQMEAI---HIASTPAELYNAVLVDTPLANYFVDCIN-------------------EQD 169
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +V+++RN+L K ++EDFY F L T EVM IL EAD R + + INS
Sbjct 170 LDEMNVEVIRNTLYKAYIEDFYKFCAGL-GGKTAEVMCDILAFEADRRSIIITINS 224
> xla:447489 atp6v0d2, MGC81907; ATPase, H+ transporting, lysosomal
38kDa, V0 subunit d2 (EC:3.6.3.14); K02146 V-type H+-transporting
ATPase subunit AC39 [EC:3.6.3.14]
Length=288
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 61/176 (34%), Positives = 101/176 (57%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ KL EF Y ++ A++PLA FLD+I MIDN++ LI G L+++P EL+ + PLG
Sbjct 74 KEKLMTEFHYFRNHAFEPLATFLDFITYSYMIDNIILLITGTLHQRPISELVPKCHPLGS 133
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M A+ + + + EL+ ++++DTP+ +F L+ E+
Sbjct 134 FEQMEAV---NIAQTPAELFNAIIVDTPLADFFQDCLS-------------------END 171
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
M E +++++RN L K +LE FY F + L TT+E++ I++ EAD R + INS
Sbjct 172 MDEMNIEIMRNKLYKSYLEGFYKFCKKL-GGTTEEIVCPIIEFEADRRAFIITINS 226
> dre:322811 atp6v0d1, fb73h07, wu:fb73h07, zgc:63769; ATPase,
H+ transporting, V0 subunit D isoform 1 (EC:3.6.3.14); K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 61/176 (34%), Positives = 101/176 (57%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ K+ EFR++++Q+Y+PLA F+D+I MIDN++ LI G L+++ EL+ + PLG
Sbjct 74 KEKMVVEFRHMRNQSYEPLASFMDFITYSYMIDNVILLITGTLHQRAISELVPKCHPLGS 133
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M A+ + + + ELY ++L+DTP+ +F ++ E
Sbjct 134 FEQMEAV---NIAQTPAELYNAILVDTPLAAFFQDCIS-------------------EQD 171
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++++RN+L K +LE FY F TL TT + M IL+ EAD R + INS
Sbjct 172 LDEMNIEIIRNTLYKAYLEAFYKFCTTL-GGTTADTMCPILEFEADRRAFIITINS 226
> xla:379892 atp6v0d1, MGC53957, atp6d; ATPase, H+ transporting,
lysosomal 38kDa, V0 subunit d1 (EC:3.6.3.14); K02146 V-type
H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 60/176 (34%), Positives = 102/176 (57%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ K+ EFR++++Q+Y+PLA F+D+I MIDN++ LI G L+++ EL+ + PLG
Sbjct 75 KEKMVVEFRHMRNQSYEPLASFMDFITYSYMIDNVILLITGTLHQRSISELVPKCHPLGS 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M A+ + + + ELY ++L+DTP+ +F ++ E
Sbjct 135 FEQMEAV---NIAQTPAELYNAILVDTPLAAFFQDCIS-------------------EQD 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++++RN+L K +LE FY F ++L TT + M IL+ EAD R + INS
Sbjct 173 LDEMNIEIIRNTLYKAYLESFYKFCKSL-GGTTGDAMCPILEFEADRRAFIITINS 227
> mmu:11972 Atp6v0d1, AI267038, Ac39, Atp6d, P39, VATX, Vma6;
ATPase, H+ transporting, lysosomal V0 subunit D1 (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 62/176 (35%), Positives = 100/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ K+ EFR++++ AY+PLA FLD+I MIDN++ LI G L+++ EL+ + PLG
Sbjct 75 KEKMVVEFRHMRNHAYEPLASFLDFITYSYMIDNVILLITGTLHQRSIAELVPKCHPLGS 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M A+ + + + ELY ++L+DTP+ +F ++ E
Sbjct 135 FEQMEAV---NIAQTPAELYNAILVDTPLAAFFQDCIS-------------------EQD 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++++RN+L K +LE FY F TL TT + M IL+ EAD R + INS
Sbjct 173 LDEMNIEIIRNTLYKAYLESFYKFC-TLLGGTTADAMCPILEFEADRRAFIITINS 227
> hsa:9114 ATP6V0D1, ATP6D, ATP6DV, FLJ43534, P39, VATX, VMA6,
VPATPD; ATPase, H+ transporting, lysosomal 38kDa, V0 subunit
d1 (EC:3.6.3.14); K02146 V-type H+-transporting ATPase subunit
AC39 [EC:3.6.3.14]
Length=351
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 62/176 (35%), Positives = 100/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ K+ EFR++++ AY+PLA FLD+I MIDN++ LI G L+++ EL+ + PLG
Sbjct 75 KEKMVVEFRHMRNHAYEPLASFLDFITYSYMIDNVILLITGTLHQRSIAELVPKCHPLGS 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M A+ + + + ELY ++L+DTP+ +F ++ E
Sbjct 135 FEQMEAV---NIAQTPAELYNAILVDTPLAAFFQDCIS-------------------EQD 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++++RN+L K +LE FY F TL TT + M IL+ EAD R + INS
Sbjct 173 LDEMNIEIIRNTLYKAYLESFYKFC-TLLGGTTADAMCPILEFEADRRAFIITINS 227
> mmu:242341 Atp6v0d2, 1620401A02Rik, AI324824, V-ATPase; ATPase,
H+ transporting, lysosomal V0 subunit D2 (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 109 bits (273), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 61/176 (34%), Positives = 100/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
R KL EF Y ++ + +PL+ FL Y+ MIDN++ L+ GAL +K +E+LA+ PLG
Sbjct 75 RKKLCREFDYFRNHSLEPLSTFLTYMTCSYMIDNIILLMNGALQKKSVKEVLAKCHPLGR 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F EM A+ + + + +L++++L++TP+ +F ++ E+
Sbjct 135 FTEMEAV---NIAETPSDLFKAVLVETPLAPFFQDCMS-------------------ENT 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++LLRN L K +LE FY F + D T +VM IL+ EAD R L + +NS
Sbjct 173 LDELNIELLRNKLYKSYLEAFYKFCKDHGD-VTADVMCPILEFEADRRALIITLNS 227
> hsa:245972 ATP6V0D2, ATP6D2, FLJ38708, VMA6; ATPase, H+ transporting,
lysosomal 38kDa, V0 subunit d2 (EC:3.6.3.14); K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 108 bits (270), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 60/176 (34%), Positives = 96/176 (54%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
R +L EF Y ++ + +PL+ FL Y+ MIDN++ L+ GAL +K +E+L + PLG
Sbjct 75 RKRLCGEFEYFRNHSLEPLSTFLTYMTCSYMIDNVILLMNGALQKKSVKEILGKCHPLGR 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F EM A+ + + + +L+ ++LI+TP+ +F ++ A
Sbjct 135 FTEMEAV---NIAETPSDLFNAILIETPLAPFFQDCMSENA------------------- 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++LLRN L K +LE FY F + D T EVM IL+ EAD R + +NS
Sbjct 173 LDELNIELLRNKLYKSYLEAFYKFCKNHGD-VTAEVMCPILEFEADRRAFIITLNS 227
> ath:AT3G28715 H+-transporting two-sector ATPase, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 105 bits (263), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 100/174 (57%), Gaps = 23/174 (13%)
Query 4 KLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGYFG 63
KL +++++ QA +P++ FL+YI MIDN++ ++ G L+ + +EL+ + PLG F
Sbjct 77 KLVDDYKHMLCQATEPMSTFLEYIRYGHMIDNVVLIVTGTLHERDVQELIEKCHPLGMFD 136
Query 64 EMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHIMT 123
+IA + + + ELYR +L+DTP+ YF + T ++ +
Sbjct 137 ---SIATLAVAQNMRELYRLVLVDTPLAPYF-------SECLTSED------------LD 174
Query 124 EADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ +++++RN+L K +LEDFY F Q L T E+M+ +L EAD R +++ INS
Sbjct 175 DMNIEIMRNTLYKAYLEDFYNFCQKL-GGATAEIMSDLLAFEADRRAVNITINS 227
> ath:AT3G28710 H+-transporting two-sector ATPase, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 105 bits (262), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 100/174 (57%), Gaps = 23/174 (13%)
Query 4 KLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGYFG 63
KL +++++ QA +P++ FL+YI MIDN++ ++ G L+ + +EL+ + PLG F
Sbjct 77 KLVDDYKHMLCQATEPMSTFLEYIRYGHMIDNVVLIVTGTLHERDVQELIEKCHPLGMFD 136
Query 64 EMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHIMT 123
+IA + + + ELYR +L+DTP+ YF + T ++ +
Sbjct 137 ---SIATLAVAQNMRELYRLVLVDTPLAPYF-------SECLTSED------------LD 174
Query 124 EADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ +++++RN+L K +LEDFY F Q L T E+M+ +L EAD R +++ INS
Sbjct 175 DMNIEIMRNTLYKAYLEDFYKFCQKL-GGATAEIMSDLLAFEADRRAVNITINS 227
> sce:YLR447C VMA6; Subunit d of the five-subunit V0 integral
membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic
proton pump found in the endomembrane system; stabilizes
VO subunits; required for V1 domain assembly on the vacuolar
membrane (EC:3.6.3.14); K02146 V-type H+-transporting ATPase
subunit AC39 [EC:3.6.3.14]
Length=345
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 59/177 (33%), Positives = 92/177 (51%), Gaps = 21/177 (11%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
A SKL EF Y++ Q+ KF+DYI MIDN+ +I G ++ + E+L R PLG
Sbjct 72 ASSKLYHEFNYIRDQSSGSTRKFMDYITYGYMIDNVALMITGTIHDRDKGEILQRCHPLG 131
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
+F + ++ ++ E LY ++L+DTP+ YF N A
Sbjct 132 WFDTLPTLS---VATDLESLYETVLVDTPLAPYF----KNCFDTAEE------------- 171
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ + +++++RN L K +LEDFY FV KE M +L EAD R +++A+NS
Sbjct 172 -LDDMNIEIIRNKLYKAYLEDFYNFVTEEIPEPAKECMQTLLGFEADRRSINIALNS 227
> ath:AT4G24840 hypothetical protein
Length=756
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 54/114 (47%), Gaps = 6/114 (5%)
Query 53 LARADPLGYFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTG 112
L +D + +RA A +D +++ EE++R+ ++ P + T AA T ++
Sbjct 256 LNNSDTSVLYNCLRAYAAIDNTNAAEEIFRTTIV-APFIQKIITHETTTNAAGTSEDELE 314
Query 113 HSLRQIEHIMTEADVDLLR-NSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTE 165
+ +QI+H + + LL +S K L F L +S KEV+ I K +
Sbjct 315 NDYKQIKHFIAKDCKMLLEISSTDKSGLHVF----DFLANSILKEVLWAIQKVK 364
Lambda K H
0.320 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40