bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1508_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
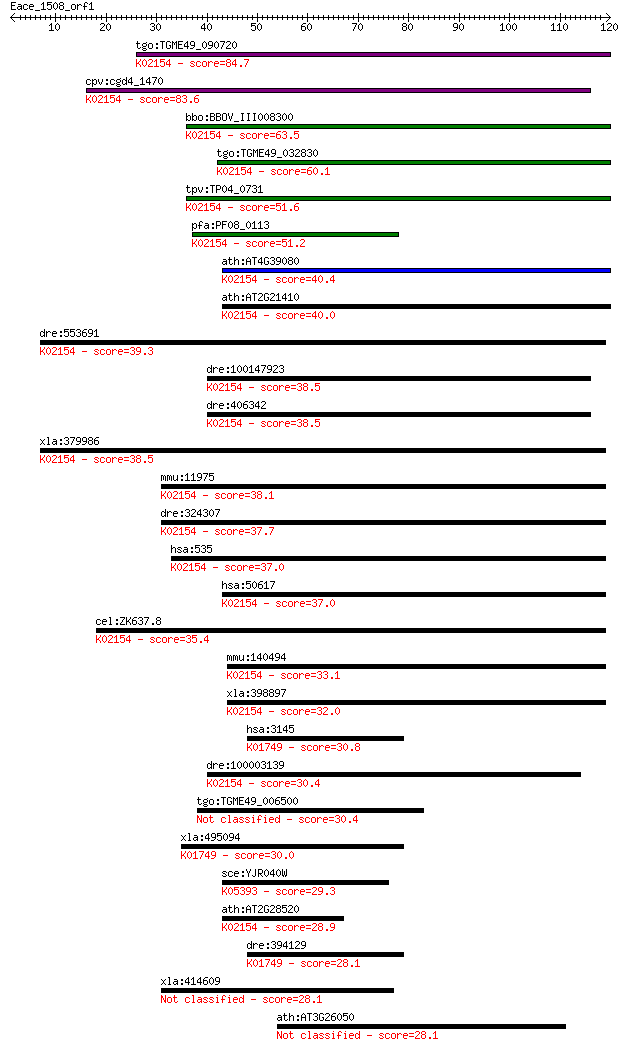
tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit... 84.7 7e-17
cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 tran... 83.6 1e-16
bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit fam... 63.5 2e-10
tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit... 60.1 2e-09
tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14); K... 51.6 6e-07
pfa:PF08_0113 vacuolar proton translocating ATPase subunit A, ... 51.2 8e-07
ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPa... 40.4 0.001
ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPa... 40.0 0.002
dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPas... 39.3 0.003
dre:100147923 T-cell immune regulator 1-like; K02154 V-type H+... 38.5 0.005
dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154 V... 38.5 0.005
xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysoso... 38.5 0.006
mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1... 38.1 0.006
dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,... 37.7 0.009
hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1, ... 37.0 0.014
hsa:50617 ATP6V0A4, A4, ATP6N1B, ATP6N2, MGC130016, MGC130017,... 37.0 0.017
cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K021... 35.4 0.048
mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lys... 33.1 0.20
xla:398897 atp6v0a4, MGC68661; ATPase, H+ transporting, lysoso... 32.0 0.45
hsa:3145 HMBS, PBG-D, PBGD, PORC, UPS; hydroxymethylbilane syn... 30.8 1.2
dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator... 30.4 1.3
tgo:TGME49_006500 hypothetical protein 30.4 1.3
xla:495094 hmbs.2, pbg-d, pbgd, ups; hydroxymethylbilane synth... 30.0 1.8
sce:YJR040W GEF1, CLC; Gef1p; K05393 chloride channel, other e... 29.3 3.5
ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATP... 28.9 4.4
dre:394129 hmbsa, MGC64128, hmbs, zgc:64128; hydroxymethylbila... 28.1 6.5
xla:414609 iqcb1, MGC81507, nphp5, piq, slsn5; IQ motif contai... 28.1 6.8
ath:AT3G26050 hypothetical protein 28.1 7.4
> tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=1015
Score = 84.7 bits (208), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 48/96 (50%), Positives = 64/96 (66%), Gaps = 2/96 (2%)
Query 26 GLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTA-YPH 84
G G E S+ FSSVAG++ + DQE+FARA+FRS RGNA+T+F+ + E+ A Y
Sbjct 241 GFGDELGSEFRSMAFSSVAGVINSCDQEKFARALFRSMRGNAYTYFQPADLTEVPAEYAV 300
Query 85 GEEPKSLFVTYFQGAAT-GSAFMEKVKCVCSAMGAR 119
+ KSLFVTY+QG + SA EKV +C+A GAR
Sbjct 301 TLQSKSLFVTYYQGGCSPSSAAYEKVIRLCAAFGAR 336
> cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 transmembrane
regions near C-terminus ; K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=920
Score = 83.6 bits (205), Expect = 1e-16, Method: Composition-based stats.
Identities = 42/101 (41%), Positives = 64/101 (63%), Gaps = 2/101 (1%)
Query 16 SASRRLLSAGGLGGLES-GEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRL 74
+ S L++ G + G+ + ++FSS+AG+V +DQE+FARA+FR+TRGN FTHF+ +
Sbjct 184 TPSSPLMNPGIMDGINNVSGFGDMMFSSIAGVVKHEDQEKFARALFRATRGNTFTHFQSI 243
Query 75 EEEELTAYPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSA 115
E + + K +FV YFQGA T SA +K+ +C A
Sbjct 244 AENIMDPKTSKDVQKVVFVIYFQGATT-SAVYDKISRICDA 283
> bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit family
protein; K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=927
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 35/97 (36%), Positives = 60/97 (61%), Gaps = 16/97 (16%)
Query 36 SSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKR-------------LEEEELTAY 82
S+L FS++AG+++A+D++ F+RA+FR+ RGN +T F+ + EEE A
Sbjct 199 STLAFSNIAGVISAEDKDAFSRAIFRAMRGNVYTFFQDSQVIKEAILSRGLITEEE--AS 256
Query 83 PHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGAR 119
G E K +FV Y Q +A+GS+ +K++ +C+ A+
Sbjct 257 IRGNEEKIVFVIYCQ-SASGSSTFQKLQKLCNGFQAK 292
> tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=909
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 47/78 (60%), Gaps = 1/78 (1%)
Query 42 SVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAAT 101
+VAG++A RF R +FR+TRGNAF F+ E+ + + E K +FV Y+QGAA
Sbjct 213 TVAGMIATSAIGRFQRMLFRTTRGNAFCFFQSTAEKLVDSNTGNEVEKGVFVIYYQGAA- 271
Query 102 GSAFMEKVKCVCSAMGAR 119
S EK+ VC+A A+
Sbjct 272 HSLLREKIVKVCAAFDAK 289
> tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=936
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 55/97 (56%), Gaps = 18/97 (18%)
Query 36 SSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKR-------------LEEEELTAY 82
SS+ F++++G+++++++E F+RA+FR+ RGN FT +++EEL
Sbjct 197 SSISFTNISGLISSQEKEAFSRAIFRAMRGNVFTLLHDTNDLRSMVLSKGLVDQEELDT- 255
Query 83 PHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGAR 119
+ K++FV Y Q ++ A K+K +C+ A+
Sbjct 256 ---DNDKTVFVIYCQ-SSNNDATYNKIKKLCTGFQAK 288
> pfa:PF08_0113 vacuolar proton translocating ATPase subunit A,
putative; K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=1053
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 34/41 (82%), Gaps = 0/41 (0%)
Query 37 SLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEE 77
+++F++++G++ KDQE F+R +FR+ RGN +T+F+ ++E+
Sbjct 198 NMMFTNISGVIKTKDQESFSRTIFRAFRGNTYTYFQNIDED 238
> ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 4/77 (5%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATG 102
+ G+V + F R +FR+TRGN F +EE + + K++FV ++ G
Sbjct 194 LTGLVPREKSMVFERILFRATRGNIFIRQTVIEEPVIDPNSGEKAEKNVFVVFYSGERAK 253
Query 103 SAFMEKVKCVCSAMGAR 119
S K+ +C A GA
Sbjct 254 S----KILKICEAFGAN 266
> ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 4/77 (5%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATG 102
+ G+V + F R +FR+TRGN F +EE + + K++FV ++ G
Sbjct 195 LTGLVPREKSMVFERILFRATRGNIFIRQSVIEESVVDPNSGEKAEKNVFVVFYSGERAK 254
Query 103 SAFMEKVKCVCSAMGAR 119
S K+ +C A GA
Sbjct 255 S----KILKICEAFGAN 267
> dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1b; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 48/114 (42%), Gaps = 6/114 (5%)
Query 7 QEKQQTFPYSASRRLL--SAGGLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTR 64
+ QQ F LL S+ L E+G + L VAG++ + F R ++R R
Sbjct 133 RRTQQFFDEMEDPNLLEESSSLLDPSEAGRGAPLRLGFVAGVINRERIPTFERMLWRVCR 192
Query 65 GNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
GN F +E+ + KS+F+ +FQ G +VK +C A
Sbjct 193 GNVFLRQTEIEDPLEDPTTGDQVHKSVFIIFFQ----GDQLKNRVKKICEGFRA 242
> dre:100147923 T-cell immune regulator 1-like; K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=793
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 37/78 (47%), Gaps = 8/78 (10%)
Query 40 FSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEE--PKSLFVTYFQ 97
S VAG+V F R ++R+ RG F +EE+ +PH +E ++F+ F
Sbjct 163 LSFVAGVVHPWKVPAFERLLWRACRGYIIVDFHEMEEK--LEHPHTDEQLQWTVFLISFW 220
Query 98 GAATGSAFMEKVKCVCSA 115
G G +KVK +C
Sbjct 221 GDQIG----QKVKKICDC 234
> dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 37/78 (47%), Gaps = 8/78 (10%)
Query 40 FSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEE--PKSLFVTYFQ 97
S VAG+V F R ++R+ RG F +EE+ +PH +E ++F+ F
Sbjct 163 LSFVAGVVHPWKVPAFERLLWRACRGYIIVDFHEMEEK--LEHPHTDEQLQWTVFLISFW 220
Query 98 GAATGSAFMEKVKCVCSA 115
G G +KVK +C
Sbjct 221 GDQIG----QKVKKICDC 234
> xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysosomal
V0 subunit a1 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=831
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 48/114 (42%), Gaps = 6/114 (5%)
Query 7 QEKQQTFPYSASRRLL--SAGGLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTR 64
++ QQ F A LL S+ L E G + L VAG++ + F R ++R R
Sbjct 133 RKTQQFFDEMADPDLLEESSTLLEPSEMGRGAPLRLGFVAGVINRERIPTFERMLWRVCR 192
Query 65 GNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
GN F ++E KS+F+ +FQ G +VK +C A
Sbjct 193 GNVFLRQAQIENPLEDPVTGDSVHKSVFIIFFQ----GDQLKNRVKKICEGFRA 242
> mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1,
Vpp-1, Vpp1; ATPase, H+ transporting, lysosomal V0 subunit
A1 (EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 42/92 (45%), Gaps = 12/92 (13%)
Query 31 ESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLE---EEELTA-YPHGE 86
E G + L VAG++ + F R ++R RGN F +E E+ +T Y H
Sbjct 159 EMGRGAPLRLGFVAGVINRERIPTFERMLWRVCRGNVFLRQAEIENPLEDPVTGDYVH-- 216
Query 87 EPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
KS+F+ +FQ G +VK +C A
Sbjct 217 --KSVFIIFFQ----GDQLKNRVKKICEGFRA 242
> dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1a (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 37/88 (42%), Gaps = 4/88 (4%)
Query 31 ESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKS 90
E G + L VAG++ + F R ++R RGN F +E+ KS
Sbjct 159 EVGRAAPLRLGFVAGVIGRERIPTFERMLWRVCRGNVFLRQADIEDPLEDPATGDNVHKS 218
Query 91 LFVTYFQGAATGSAFMEKVKCVCSAMGA 118
+F+ +FQ G +VK +C A
Sbjct 219 VFIIFFQ----GDQLKNRVKKICEGFRA 242
> hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1,
Vph1, a1; ATPase, H+ transporting, lysosomal V0 subunit a1
(EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 41/90 (45%), Gaps = 12/90 (13%)
Query 33 GEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLE---EEELTA-YPHGEEP 88
G + L VAG++ + F R ++R RGN F +E E+ +T Y H
Sbjct 168 GRGTPLRLGFVAGVINRERIPTFERMLWRVCRGNVFLRQAEIENPLEDPVTGDYVH---- 223
Query 89 KSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
KS+F+ +FQ G +VK +C A
Sbjct 224 KSVFIIFFQ----GDQLKNRVKKICEGFRA 249
> hsa:50617 ATP6V0A4, A4, ATP6N1B, ATP6N2, MGC130016, MGC130017,
RDRTA2, RTA1C, RTADR, STV1, VPH1, VPP2; ATPase, H+ transporting,
lysosomal V0 subunit a4 (EC:3.6.3.14); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=840
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 18/76 (23%), Positives = 35/76 (46%), Gaps = 4/76 (5%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATG 102
+AG++ + F R ++R RGN + F ++ E K++F+ ++Q G
Sbjct 173 IAGVINRERMASFERLLWRICRGNVYLKFSEMDAPLEDPVTKEEIQKNIFIIFYQ----G 228
Query 103 SAFMEKVKCVCSAMGA 118
+K+K +C A
Sbjct 229 EQLRQKIKKICDGFRA 244
> cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=899
Score = 35.4 bits (80), Expect = 0.048, Method: Composition-based stats.
Identities = 28/108 (25%), Positives = 48/108 (44%), Gaps = 14/108 (12%)
Query 18 SRRLLSAGGLGGLESGEMSS----LVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKR 73
+R L+ G + + MS+ L VAG++ + F R ++R+ RGN F
Sbjct 178 TRPLIDIGDMDDDSAARMSAQAAMLRLGFVAGVIQRERLPAFERLLWRACRGNVFLRTSE 237
Query 74 LEE---EELTAYPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
+++ + +T P K +F+ +FQ G KVK +C A
Sbjct 238 IDDVLNDTVTGDPVN---KCVFIIFFQ----GDHLKTKVKKICEGFRA 278
> mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lysosomal
V0 subunit A4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=833
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 33/75 (44%), Gaps = 4/75 (5%)
Query 44 AGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATGS 103
AG++ + F R ++R RGN + F ++ E K++F+ ++Q G
Sbjct 174 AGVINRERMASFERLLWRVCRGNVYLKFSEMDTLLEDPVTKEEIKKNIFIIFYQ----GE 229
Query 104 AFMEKVKCVCSAMGA 118
K+K +C A
Sbjct 230 QLRLKIKKICDGFRA 244
> xla:398897 atp6v0a4, MGC68661; ATPase, H+ transporting, lysosomal
V0 subunit a4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=846
Score = 32.0 bits (71), Expect = 0.45, Method: Composition-based stats.
Identities = 17/75 (22%), Positives = 33/75 (44%), Gaps = 4/75 (5%)
Query 44 AGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATGS 103
AG++ + F R ++R RGN + + ++ E K++F+ ++Q G
Sbjct 176 AGVINRERMATFERLLWRVCRGNIYLKYTEMDMALEDPITKEEVKKNVFIIFYQ----GD 231
Query 104 AFMEKVKCVCSAMGA 118
K+K +C A
Sbjct 232 QLKLKIKKICDGFKA 246
> hsa:3145 HMBS, PBG-D, PBGD, PORC, UPS; hydroxymethylbilane synthase
(EC:2.5.1.61); K01749 hydroxymethylbilane synthase [EC:2.5.1.61]
Length=361
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 48 AAKDQERFARAMFRSTRGNAFTHFKRLEEEE 78
AA+ Q +F FRS RGN T ++L+E++
Sbjct 151 AAQLQRKFPHLEFRSIRGNLNTRLRKLDEQQ 181
> dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator
1, ATPase, H+ transporting, lysosomal V0 subunit A3; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query 40 FSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPK-SLFVTYFQG 98
S VAG+V F R ++R+ RG F + EE L GE + ++FV F G
Sbjct 167 LSFVAGVVHPWKVPAFERLLWRACRGYIIVDFWEM-EERLEEPDTGETIQWTVFVISFWG 225
Query 99 AATGSAFMEKVKCVC 113
G +KVK +C
Sbjct 226 EQIG----QKVKKIC 236
> tgo:TGME49_006500 hypothetical protein
Length=1102
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 27/45 (60%), Gaps = 3/45 (6%)
Query 38 LVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAY 82
L++ ++A++D RA++ T+G+AF H+ RL+ E + +
Sbjct 1016 LLYERFGHLIASRDD---WRAVYVLTKGSAFQHYSRLDWERVLTW 1057
> xla:495094 hmbs.2, pbg-d, pbgd, ups; hydroxymethylbilane synthase
(EC:2.5.1.61); K01749 hydroxymethylbilane synthase [EC:2.5.1.61]
Length=256
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 4/48 (8%)
Query 35 MSSLVFSSVAGIV----AAKDQERFARAMFRSTRGNAFTHFKRLEEEE 78
+S+L SV G AA+ +++F F+ RGN T K+L+E+E
Sbjct 29 LSTLPEKSVIGTSSLRRAAQLKKKFPHLEFKDIRGNLNTRLKKLDEQE 76
> sce:YJR040W GEF1, CLC; Gef1p; K05393 chloride channel, other
eukaryote
Length=779
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLE 75
+ G+V AKD RF R +R G FT+ + L+
Sbjct 724 LKGLVTAKDILRFKRIKYREVHGAKFTYNEALD 756
> ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=817
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 43 VAGIVAAKDQERFARAMFRSTRGN 66
++GI+ +F R +FR+TRGN
Sbjct 193 ISGIINKDKLLKFERMLFRATRGN 216
> dre:394129 hmbsa, MGC64128, hmbs, zgc:64128; hydroxymethylbilane
synthase a; K01749 hydroxymethylbilane synthase [EC:2.5.1.61]
Length=358
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 48 AAKDQERFARAMFRSTRGNAFTHFKRLEEEE 78
AA+ ++RF + F + RGN T K+L+E++
Sbjct 149 AAQLKKRFPQLEFENIRGNLNTRLKKLDEKD 179
> xla:414609 iqcb1, MGC81507, nphp5, piq, slsn5; IQ motif containing
B1
Length=594
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 31 ESGEMSSLVFSSVAG---IVAAKDQERFARAMFRSTRGNAFTHFKRLEE 76
E+ E S + S G ++A + Q R+ RAM RG F+ F+R+ +
Sbjct 98 EAEEFYSKLLPSALGNMLLLARRIQARYVRAMKEEDRGELFSSFRRVSD 146
> ath:AT3G26050 hypothetical protein
Length=533
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 54 RFARAMFRSTRGNAFTHFKRLEEEELTAYPHG-EEPKSLFVTYFQGAATGSAFMEKVK 110
R+A R +AFT + LEE E P E K+ F +F+ A+G A + K
Sbjct 39 RYANETLAWARWSAFTQNRYLEEVERFTKPGSVAEKKAFFEAHFKNRASGKAATQTKK 96
Lambda K H
0.317 0.129 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022937320
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40