bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
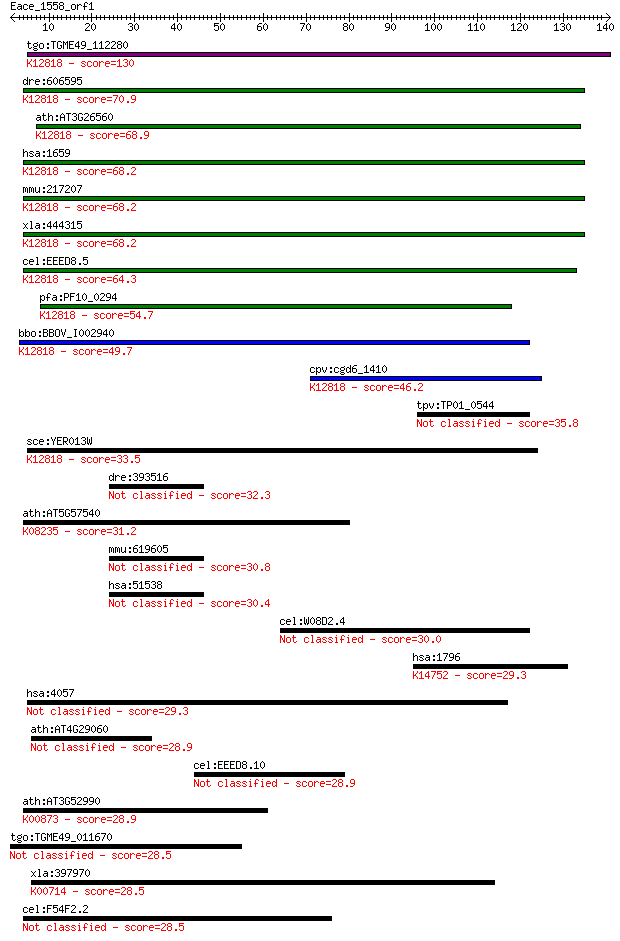
Query= Eace_1558_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112280 ATP-dependent RNA helicase, putative (EC:3.4... 130 9e-31
dre:606595 im:7153552; K12818 ATP-dependent RNA helicase DHX8/... 70.9 1e-12
ath:AT3G26560 ATP-dependent RNA helicase, putative; K12818 ATP... 68.9 5e-12
hsa:1659 DHX8, DDX8, HRH1, PRP22, PRPF22; DEAH (Asp-Glu-Ala-Hi... 68.2 7e-12
mmu:217207 Dhx8, Ddx8, KIAA4096, MGC31290, mDEAH6, mKIAA4096; ... 68.2 7e-12
xla:444315 MGC80994 protein; K12818 ATP-dependent RNA helicase... 68.2 9e-12
cel:EEED8.5 mog-5; Masculinisation Of Germline family member (... 64.3 1e-10
pfa:PF10_0294 RNA helicase, putative; K12818 ATP-dependent RNA... 54.7 1e-07
bbo:BBOV_I002940 19.m02117; RNA helicase; K12818 ATP-dependent... 49.7 3e-06
cpv:cgd6_1410 pre-mRNA splicing factor ATP-dependent RNA helic... 46.2 3e-05
tpv:TP01_0544 RNA helicase 35.8 0.042
sce:YER013W PRP22; DEAH-box RNA-dependent ATPase/ATP-dependent... 33.5 0.24
dre:393516 zcchc17, MGC65790, ps1d, zgc:65790, zgc:85656; zinc... 32.3 0.46
ath:AT5G57540 xyloglucan:xyloglucosyl transferase, putative / ... 31.2 1.1
mmu:619605 Zcchc17, 2810055E05Rik, HSPC251, LDC4, pNO40, pS1D;... 30.8 1.4
hsa:51538 ZCCHC17, HSPC251, PS1D, RP11-266K22.1, pNO40; zinc f... 30.4 1.8
cel:W08D2.4 fat-3; FATty acid desaturase family member (fat-3) 30.0 2.1
hsa:1796 DOK1, MGC117395, MGC138860, P62DOK; docking protein 1... 29.3 3.8
hsa:4057 LTF, GIG12, HLF2, LF; lactotransferrin 29.3 4.4
ath:AT4G29060 emb2726 (embryo defective 2726); RNA binding / t... 28.9 4.8
cel:EEED8.10 hypothetical protein 28.9 5.3
ath:AT3G52990 pyruvate kinase, putative (EC:2.7.1.40); K00873 ... 28.9 5.5
tgo:TGME49_011670 S1 RNA-binding domain containing protein (EC... 28.5 6.3
xla:397970 wnt8a, MGC80495, Xwnt8, wnt-8, wnt8, xwnt-8; wingle... 28.5 6.5
cel:F54F2.2 zfp-1; Zinc Finger Protein family member (zfp-1) 28.5 7.6
> tgo:TGME49_112280 ATP-dependent RNA helicase, putative (EC:3.4.22.44);
K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1206
Score = 130 bits (328), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 67/139 (48%), Positives = 97/139 (69%), Gaps = 7/139 (5%)
Query 5 DAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRN---LQSGNAAGSRAALFESL 61
D V + +VKVK+ IAG+++SL+MREVDQ TG DLKPR + + AG + +
Sbjct 287 DVVHRNMVVKVKILGIAGTKISLSMREVDQETGEDLKPRGEFARKEEDTAGEKKKI---R 343
Query 62 AEEHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKE 121
++ A+ EK+G+GR+TG++I+ + E+ Y RKRKLMSD+DKWEAQQL SGL++ +E
Sbjct 344 IDDEAEEREKQGIGRLTGIRIDSKDNNVESLYCRKRKLMSDFDKWEAQQLLHSGLLTREE 403
Query 122 NPYYDEEAGGLLPSQEAEE 140
+P +DEE G+LPS E +E
Sbjct 404 HPLFDEEL-GILPSAEVDE 421
> dre:606595 im:7153552; K12818 ATP-dependent RNA helicase DHX8/PRP22
[EC:3.6.4.13]
Length=1210
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/132 (37%), Positives = 70/132 (53%), Gaps = 15/132 (11%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
AD V KGQ VKVKV + GS+ SL+M++VDQ TG DL P ++ G E
Sbjct 300 ADVVSKGQRVKVKVLSFTGSKTSLSMKDVDQDTGEDLNPNRRRNVGGEGQ---------E 350
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKRKL-MSDYDKWEAQQLQRSGLVSSKEN 122
E A + R T + + E+ RKR +SD +KWE +Q+ + ++S +E
Sbjct 351 ESAM----RNPDRPTNLNLGHAPEVEDDTLERKRLTKISDPEKWEIKQMIAANVLSKEEF 406
Query 123 PYYDEEAGGLLP 134
P +DEE G+LP
Sbjct 407 PDFDEET-GILP 417
> ath:AT3G26560 ATP-dependent RNA helicase, putative; K12818 ATP-dependent
RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1168
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 50/129 (38%), Positives = 70/129 (54%), Gaps = 12/129 (9%)
Query 7 VKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAEEHA 66
VK+ V VKV +I+ + SL+MR+VDQ TGRDL P S SR+ S +
Sbjct 260 VKRDMEVYVKVISISSDKYSLSMRDVDQNTGRDLIPLRKPSDEDDSSRSN--PSYRTKDG 317
Query 67 QALEKKGLGRITGVKINIESTEEETAYARKRKL--MSDYDKWEAQQLQRSGLVSSKENPY 124
Q + K G I+G++I EE +R L MS ++WEA+QL SG++ E P
Sbjct 318 Q-VTKTG---ISGIRI----VEENDVAPSRRPLKKMSSPERWEAKQLIASGVLRVDEFPM 369
Query 125 YDEEAGGLL 133
YDE+ G+L
Sbjct 370 YDEDGDGML 378
> hsa:1659 DHX8, DDX8, HRH1, PRP22, PRPF22; DEAH (Asp-Glu-Ala-His)
box polypeptide 8 (EC:3.6.4.13); K12818 ATP-dependent RNA
helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1220
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 46/132 (34%), Positives = 70/132 (53%), Gaps = 15/132 (11%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
AD V KGQ VKVKV + G++ SL+M++VDQ TG DL P + +L
Sbjct 310 ADVVSKGQRVKVKVLSFTGTKTSLSMKDVDQETGEDLNPNRRR-------------NLVG 356
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKR-KLMSDYDKWEAQQLQRSGLVSSKEN 122
E + + R T + + E+ + RKR +SD +KWE +Q+ + ++S +E
Sbjct 357 ETNEETSMRNPDRPTHLSLVSAPEVEDDSLERKRLTRISDPEKWEIKQMIAANVLSKEEF 416
Query 123 PYYDEEAGGLLP 134
P +DEE G+LP
Sbjct 417 PDFDEET-GILP 427
> mmu:217207 Dhx8, Ddx8, KIAA4096, MGC31290, mDEAH6, mKIAA4096;
DEAH (Asp-Glu-Ala-His) box polypeptide 8 (EC:3.6.4.13); K12818
ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1244
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 46/132 (34%), Positives = 70/132 (53%), Gaps = 15/132 (11%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
AD V KGQ VKVKV + G++ SL+M++VDQ TG DL P + +L
Sbjct 334 ADVVSKGQRVKVKVLSFTGTKTSLSMKDVDQETGEDLNPNRRR-------------NLVG 380
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKR-KLMSDYDKWEAQQLQRSGLVSSKEN 122
E + + R T + + E+ + RKR +SD +KWE +Q+ + ++S +E
Sbjct 381 ETNEETSMRNPDRPTHLSLVSAPEVEDDSLERKRLTRISDPEKWEIKQMIAANVLSKEEF 440
Query 123 PYYDEEAGGLLP 134
P +DEE G+LP
Sbjct 441 PDFDEET-GILP 451
> xla:444315 MGC80994 protein; K12818 ATP-dependent RNA helicase
DHX8/PRP22 [EC:3.6.4.13]
Length=793
Score = 68.2 bits (165), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 71/136 (52%), Gaps = 23/136 (16%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKP---RNLQ-SGNAAGSRAALFE 59
AD V KGQ VK+KV + G + SL+M++V+Q TG DL P RNL GN S
Sbjct 266 ADVVSKGQRVKIKVLSFTGCKTSLSMKDVNQDTGEDLNPNRRRNLIGDGNEEASM----- 320
Query 60 SLAEEHAQALEKKGLGRITGVK-INIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVS 118
+ R T + +N E++T ++ +SD +KWE +Q+ + ++S
Sbjct 321 ------------RNPDRPTHLSLVNAPEVEDDTLERKRLTKISDPEKWEIKQMIAANVLS 368
Query 119 SKENPYYDEEAGGLLP 134
+E P +DEE G+LP
Sbjct 369 KEEFPDFDEET-GILP 383
> cel:EEED8.5 mog-5; Masculinisation Of Germline family member
(mog-5); K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1200
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/130 (32%), Positives = 69/130 (53%), Gaps = 11/130 (8%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQ-SGNAAGSRAALFESLA 62
AD +K+G+ VKVKV+ I ++SL+M+EVDQ +G DL PR + +A G R +
Sbjct 278 ADVLKRGENVKVKVNKIENGKISLSMKEVDQNSGEDLNPRETDLNPDAIGVRPRTPPAST 337
Query 63 EEHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKEN 122
E G+G+ I + R +S ++WE +Q+Q +G++++ +
Sbjct 338 SSWMNP-EASGVGQGPSTSI---------GGGKARVRISTPERWELRQMQGAGVLTATDM 387
Query 123 PYYDEEAGGL 132
P +DEE G L
Sbjct 388 PDFDEEMGVL 397
> pfa:PF10_0294 RNA helicase, putative; K12818 ATP-dependent RNA
helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1290
Score = 54.7 bits (130), Expect = 1e-07, Method: Composition-based stats.
Identities = 39/110 (35%), Positives = 56/110 (50%), Gaps = 9/110 (8%)
Query 8 KKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAEEHAQ 67
K+ VKVKV I ++SL M EVDQ TG++L N + +LF+ + E+
Sbjct 379 KRNMKVKVKVKGIFNQKISLNMSEVDQKTGKNLV--NDDMNKEEDNYISLFDDINEDF-N 435
Query 68 ALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLV 117
L+KK + + +E Y K+ SDY KWE QQL +SG+V
Sbjct 436 DLKKK------NANVFKDDPKETLKYESVIKIQSDYSKWEIQQLIKSGVV 479
> bbo:BBOV_I002940 19.m02117; RNA helicase; K12818 ATP-dependent
RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1156
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 61/123 (49%), Gaps = 24/123 (19%)
Query 3 SADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDL---KPRNLQSGNAAGSRAALFE 59
+++A+K+ +V VK + ++SL+M+ VDQ TGRDL ++QS G A + +
Sbjct 258 ASEALKEDMVVYVKKIGMKNDKISLSMKSVDQKTGRDLCSGHQDSMQSYYTMG--APMLQ 315
Query 60 SLAEEHAQALE-KKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVS 118
+ A L+ G+ RKR++M+D ++WE QQL SG++
Sbjct 316 NTASSSDAILDYSNGMD------------------GRKRRMMTDLERWEHQQLVNSGVLP 357
Query 119 SKE 121
E
Sbjct 358 KSE 360
> cpv:cgd6_1410 pre-mRNA splicing factor ATP-dependent RNA helicase
; K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1005
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 22/56 (39%), Positives = 38/56 (67%), Gaps = 3/56 (5%)
Query 71 KKGLGRITGVKINIESTEEETAYA--RKRKLMSDYDKWEAQQLQRSGLVSSKENPY 124
KKG G I+G+K+ ++S ++ Y+ ++ ++ +DY+KWE QL SG++S E PY
Sbjct 148 KKGFGAISGIKL-VDSCNVKSQYSGNKQERVSNDYEKWEIMQLLNSGVISRDEIPY 202
> tpv:TP01_0544 RNA helicase
Length=910
Score = 35.8 bits (81), Expect = 0.042, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 22/26 (84%), Gaps = 0/26 (0%)
Query 96 KRKLMSDYDKWEAQQLQRSGLVSSKE 121
KR+ ++D ++WE QQL +SG++SS+E
Sbjct 4 KRRYINDIERWENQQLLKSGILSSEE 29
> sce:YER013W PRP22; DEAH-box RNA-dependent ATPase/ATP-dependent
RNA helicase, associates with lariat intermediates before
the second catalytic step of splicing; mediates ATP-dependent
mRNA release from the spliceosome and unwinds RNA duplexes
(EC:3.6.1.-); K12818 ATP-dependent RNA helicase DHX8/PRP22
[EC:3.6.4.13]
Length=1145
Score = 33.5 bits (75), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 55/120 (45%), Gaps = 30/120 (25%)
Query 5 DAVKKGQLVKVKVHAIAGS-RLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFESLAE 63
D V++GQ + V+V I + ++SL+M+ +DQ +G +++ RN +S
Sbjct 224 DVVRQGQHIFVEVIKIQNNGKISLSMKNIDQHSG-EIRKRNTES---------------- 266
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKENP 123
+E +G + N T KR+ ++ ++WE +QL SG S + P
Sbjct 267 -----VEDRG-------RSNDAHTSRNMKNKIKRRALTSPERWEIRQLIASGAASIDDYP 314
> dre:393516 zcchc17, MGC65790, ps1d, zgc:65790, zgc:85656; zinc
finger, CCHC domain containing 17
Length=235
Score = 32.3 bits (72), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 24 RLSLTMREVDQATGRDLKPRNL 45
+LS +M V+Q TGRDL P N+
Sbjct 85 KLSFSMNSVNQGTGRDLDPNNV 106
> ath:AT5G57540 xyloglucan:xyloglucosyl transferase, putative
/ xyloglucan endotransglycosylase, putative / endo-xyloglucan
transferase, putative; K08235 xyloglucan:xyloglucosyl transferase
[EC:2.4.1.207]
Length=284
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 41/77 (53%), Gaps = 5/77 (6%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGR-DLKPRNLQSGNAAGSRAALFESLA 62
A+ V+ GQL+ + I+GS + + G+ D+K + L +GN+AG+ A + S
Sbjct 39 ANIVESGQLLTCTLDKISGSGFQ---SKKEYLFGKIDMKMK-LVAGNSAGTVTAYYLSSK 94
Query 63 EEHAQALEKKGLGRITG 79
E ++ + LG +TG
Sbjct 95 GETWDEIDFEFLGNVTG 111
> mmu:619605 Zcchc17, 2810055E05Rik, HSPC251, LDC4, pNO40, pS1D;
zinc finger, CCHC domain containing 17
Length=241
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 24 RLSLTMREVDQATGRDLKPRNL 45
++SL+M+ V+Q TG+DL P N+
Sbjct 82 KVSLSMKVVNQGTGKDLDPNNV 103
> hsa:51538 ZCCHC17, HSPC251, PS1D, RP11-266K22.1, pNO40; zinc
finger, CCHC domain containing 17
Length=241
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 24 RLSLTMREVDQATGRDLKPRNL 45
++SL+M+ V+Q TG+DL P N+
Sbjct 82 KVSLSMKVVNQGTGKDLDPNNV 103
> cel:W08D2.4 fat-3; FATty acid desaturase family member (fat-3)
Length=443
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 34/58 (58%), Gaps = 3/58 (5%)
Query 64 EHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQLQRSGLVSSKE 121
EH + LEK+ R+ V IN+ + + + A+++K++ ++K Q+L GL+ + E
Sbjct 69 EHDEFLEKQLEKRLDKVDINVSAY--DVSVAQEKKMVESFEKLR-QKLHDDGLMKANE 123
> hsa:1796 DOK1, MGC117395, MGC138860, P62DOK; docking protein
1, 62kDa (downstream of tyrosine kinase 1); K14752 docking
protein 1
Length=342
Score = 29.3 bits (64), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 95 RKRKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAG 130
RK+ L D + QQL ++ L KE+P YDE G
Sbjct 193 RKKPLYWDLYEHAQQQLLKAKLTDPKEDPIYDEPEG 228
> hsa:4057 LTF, GIG12, HLF2, LF; lactotransferrin
Length=666
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 30/126 (23%), Positives = 54/126 (42%), Gaps = 15/126 (11%)
Query 5 DAVKKGQLVKVKVHAIAGSRLS-------LTMREVDQATGRDLKPRNLQSGNAAGSRAAL 57
D K L +V HA+ ++ +R+ + G+D P+ G+ +G + L
Sbjct 215 DKFKDCHLARVPSHAVVARSVNGKEDAIWNLLRQAQEKFGKDKSPKFQLFGSPSGQKDLL 274
Query 58 FESLAEEHAQALEKK------GLGRITGVKINIESTEEETAYARKRKLMSDYDKWEAQQL 111
F+ A ++ + G G T ++ N+ +EEE A R R + + E ++
Sbjct 275 FKDSAIGFSRVPPRIDSGLYLGSGYFTAIQ-NLRKSEEEVAARRARVVWCAVGEQELRKC 333
Query 112 -QRSGL 116
Q SGL
Sbjct 334 NQWSGL 339
> ath:AT4G29060 emb2726 (embryo defective 2726); RNA binding /
translation elongation factor
Length=709
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 6 AVKKGQLVKVKVHAIAGSRLSLTMREVD 33
+++ GQ VKV+V IA R++LTM+E D
Sbjct 294 SLQAGQEVKVRVLRIARGRVTLTMKEED 321
> cel:EEED8.10 hypothetical protein
Length=738
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 44 NLQSGNAAGSRAALFESLAEEHAQALEKKGLGRIT 78
NL S +A S+ ++F++L H Q LE +GL +T
Sbjct 10 NLGSAGSAKSQQSMFKALRRLHGQNLESRGLHDMT 44
> ath:AT3G52990 pyruvate kinase, putative (EC:2.7.1.40); K00873
pyruvate kinase [EC:2.7.1.40]
Length=474
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 0/57 (0%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQATGRDLKPRNLQSGNAAGSRAALFES 60
A AVKKG + V + GS + EVD+ G D+ + + AGS L S
Sbjct 86 AKAVKKGDTIFVGQYLFTGSETTSVWLEVDEVKGDDVICLSRNAATLAGSLFTLHSS 142
> tgo:TGME49_011670 S1 RNA-binding domain containing protein (EC:2.7.7.8)
Length=378
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 29/64 (45%), Gaps = 10/64 (15%)
Query 1 CVSA-------DAVKKGQLVKVKVHAIA-GSRLSLTMREVDQATGRDLKPRNL--QSGNA 50
C+S+ DA+ G V VKV + G + L MR + Q G D P NL G
Sbjct 164 CISSQKLDRVDDALAVGDKVWVKVTKVEEGGKYGLDMRGISQKDGADNDPNNLSRSRGTR 223
Query 51 AGSR 54
+G R
Sbjct 224 SGPR 227
> xla:397970 wnt8a, MGC80495, Xwnt8, wnt-8, wnt8, xwnt-8; wingless-type
MMTV integration site family, member 8A; K00714 wingless-type
MMTV integration site family, member 8
Length=358
Score = 28.5 bits (62), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 32/126 (25%), Positives = 53/126 (42%), Gaps = 22/126 (17%)
Query 6 AVKKGQLVKVKVHAIAGS------RLSLT-MREV--------DQATGRDLKPRNLQSGNA 50
AVK+ K H I+GS L L R++ DQA ++ R ++SGN+
Sbjct 172 AVKETMKRTCKCHGISGSCSIQTCWLQLAEFRDIGNHLKIKHDQALKLEMDKRKMRSGNS 231
Query 51 AGSRAAL---FESLAEEHAQALEKKGLGRITGVKINIESTEEETAYARKRKLMSDYDKWE 107
A +R A+ F S+A LE + + + ++ TE + L +WE
Sbjct 232 ADNRGAIADAFSSVAGSELIFLEDSPDYCLKNISLGLQGTEGRECLQSGKNL----SQWE 287
Query 108 AQQLQR 113
+ +R
Sbjct 288 RRSCKR 293
> cel:F54F2.2 zfp-1; Zinc Finger Protein family member (zfp-1)
Length=211
Score = 28.5 bits (62), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 5/74 (6%)
Query 4 ADAVKKGQLVKVKVHAIAGSRLSLTMREVDQAT--GRDLKPRNLQSGNAAGSRAALFESL 61
+ +VKKG++V K T + +++A G L P ++G S++ + L
Sbjct 99 SPSVKKGEIVAPKATVTPAVN---TRKPINEAISEGIILDPTYFKNGGNLDSKSPKNQKL 155
Query 62 AEEHAQALEKKGLG 75
AEE Q ++ GLG
Sbjct 156 AEELQQFIDNPGLG 169
Lambda K H
0.308 0.125 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40