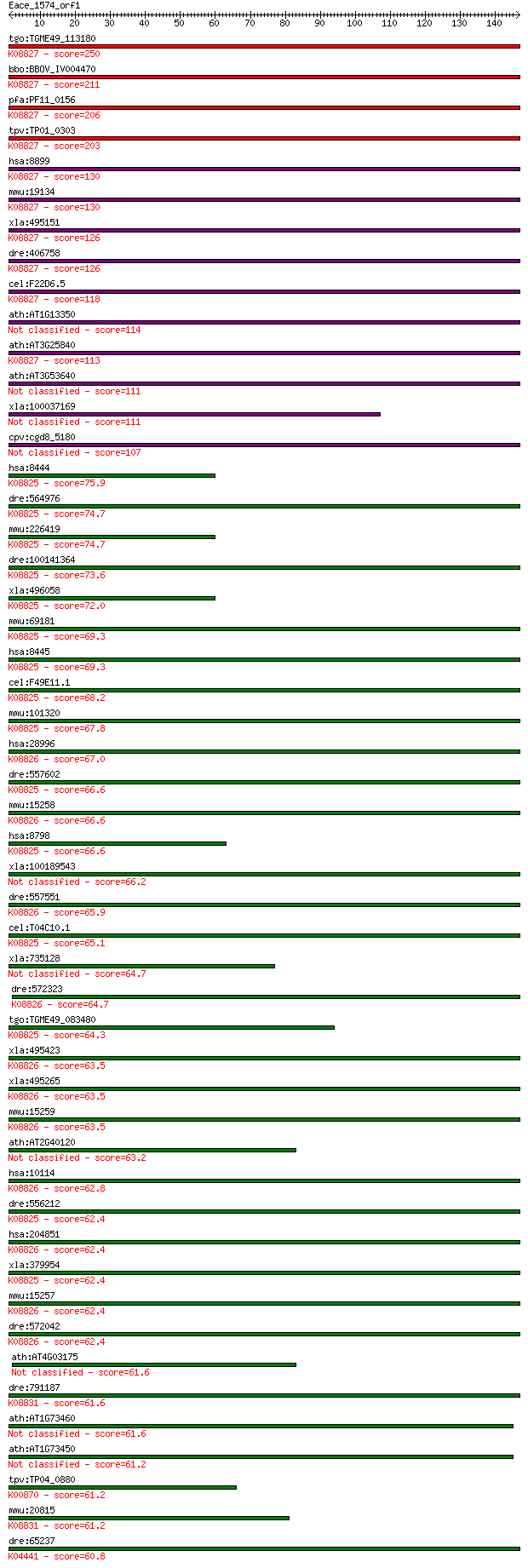
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1574_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_113180 serine/threonine-protein kinase PRP4, putati... 250 1e-66
bbo:BBOV_IV004470 23.m06285; serine/threonine-protein kinase (... 211 5e-55
pfa:PF11_0156 Ser/Thr protein kinase; K08827 serine/threonine-... 206 3e-53
tpv:TP01_0303 hypothetical protein; K08827 serine/threonine-pr... 203 2e-52
hsa:8899 PRPF4B, KIAA0536, PR4H, PRP4, PRP4H, PRP4K, dJ1013A10... 130 1e-30
mmu:19134 Prpf4b, 2610037H07, MGC5960, Prp4, Prp4k, Prpk, cbp1... 130 1e-30
xla:495151 prpf4b; PRP4 pre-mRNA processing factor 4 homolog B... 126 2e-29
dre:406758 prpf4b, wu:fc18e10, wu:fe47g02, zgc:55623; PRP4 pre... 126 3e-29
cel:F22D6.5 prpf-4; vertebrate Pre-mRNA Processing Factor fami... 118 7e-27
ath:AT1G13350 protein kinase family protein 114 9e-26
ath:AT3G25840 protein kinase family protein; K08827 serine/thr... 113 2e-25
ath:AT3G53640 protein kinase family protein 111 9e-25
xla:100037169 hypothetical protein LOC100037169 111 9e-25
cpv:cgd8_5180 hypothetical protein 107 9e-24
hsa:8444 DYRK3, DYRK5, RED, REDK, hYAK3-2; dual-specificity ty... 75.9 4e-14
dre:564976 dyrk4; dual-specificity tyrosine-(Y)-phosphorylatio... 74.7 1e-13
mmu:226419 Dyrk3, BC006704; dual-specificity tyrosine-(Y)-phos... 74.7 1e-13
dre:100141364 zgc:172180; K08825 dual-specificity tyrosine-(Y)... 73.6 2e-13
xla:496058 dyrk3; dual-specificity tyrosine-(Y)-phosphorylatio... 72.0 6e-13
mmu:69181 Dyrk2, 1810038L18Rik; dual-specificity tyrosine-(Y)-... 69.3 4e-12
hsa:8445 DYRK2, FLJ21217, FLJ21365; dual-specificity tyrosine-... 69.3 4e-12
cel:F49E11.1 mbk-2; MiniBrain Kinase (Drosophila) homolog fami... 68.2 8e-12
mmu:101320 Dyrk4, AW049118, Dyrk4a, Dyrk4b; dual-specificity t... 67.8 1e-11
hsa:28996 HIPK2, DKFZp686K02111, FLJ23711, PRO0593; homeodomai... 67.0 2e-11
dre:557602 dyrk2, fc18c09, si:ch211-266a5.8, wu:fa09g03, wu:fc... 66.6 2e-11
mmu:15258 Hipk2, 1110014O20Rik, B230339E18Rik, Stank; homeodom... 66.6 2e-11
hsa:8798 DYRK4; dual-specificity tyrosine-(Y)-phosphorylation ... 66.6 3e-11
xla:100189543 hipk1; homeodomain interacting protein kinase 1 ... 66.2 3e-11
dre:557551 hipk2, MGC198433, cb1009, si:dkey-16j24.2, si:dkey-... 65.9 4e-11
cel:T04C10.1 mbk-1; MiniBrain Kinase (Drosophila) homolog fami... 65.1 8e-11
xla:735128 MGC115587; serine/threonine-protein kinase SRPK1-like 64.7 1e-10
dre:572323 homeodomain-interacting protein kinase 3-like; K088... 64.7 1e-10
tgo:TGME49_083480 dual-specificity tyrosine-phosphorylation-re... 64.3 1e-10
xla:495423 hipk1; homeodomain interacting protein kinase 1 (EC... 63.5 2e-10
xla:495265 hipk2; homeodomain interacting protein kinase 2 (EC... 63.5 2e-10
mmu:15259 Hipk3, DYRK6, FIST3, PKY; homeodomain interacting pr... 63.5 2e-10
ath:AT2G40120 protein kinase family protein 63.2 3e-10
hsa:10114 HIPK3, DYRK6, FIST3, PKY, YAK1; homeodomain interact... 62.8 3e-10
dre:556212 dyrk1b, si:ch211-225p5.1; dual-specificity tyrosine... 62.4 4e-10
hsa:204851 HIPK1, KIAA0630, MGC26642, MGC33446, MGC33548, Myak... 62.4 4e-10
xla:379954 dyrk1a.2, MGC52802; dual-specificity tyrosine-(Y)-p... 62.4 4e-10
mmu:15257 Hipk1, 1110062K04Rik, Myak; homeodomain interacting ... 62.4 4e-10
dre:572042 homeodomain-interacting protein kinase 3-like; K088... 62.4 5e-10
ath:AT4G03175 protein kinase family protein 61.6 7e-10
dre:791187 srpk1b, zgc:158800; serine/arginine-rich protein sp... 61.6 9e-10
ath:AT1G73460 protein kinase family protein 61.6 9e-10
ath:AT1G73450 protein kinase, putative 61.2 1e-09
tpv:TP04_0880 protein kinase (EC:2.7.1.37); K00870 protein kin... 61.2 1e-09
mmu:20815 Srpk1, AU017960; serine/arginine-rich protein specif... 61.2 1e-09
dre:65237 mapk14a, MAPK14, fk28c03, hm:zeh1243, p38, p38a, wu:... 60.8 1e-09
> tgo:TGME49_113180 serine/threonine-protein kinase PRP4, putative
(EC:2.7.12.1); K08827 serine/threonine-protein kinase PRP4
[EC:2.7.11.1]
Length=921
Score = 250 bits (638), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 113/146 (77%), Positives = 133/146 (91%), Gaps = 0/146 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG +YD QIDVWSAAAT+YELATGQVLFPGRTNNDML+CIME KGK P K+I+
Sbjct 728 YRAPEIILGCRYDLQIDVWSAAATIYELATGQVLFPGRTNNDMLKCIMEVKGKIPTKMIK 787
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
AG LSS HF E+LDFI+R +D+F K++ TR+LHDLRPT+++T++L+EKQ WLKG+SPK+N
Sbjct 788 AGQLSSHHFDENLDFIYRDRDAFFKKEVTRVLHDLRPTRNLTENLIEKQHWLKGNSPKIN 847
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
FLRRKMRQLGDLLEKCL LDPQKRLT
Sbjct 848 FLRRKMRQLGDLLEKCLALDPQKRLT 873
> bbo:BBOV_IV004470 23.m06285; serine/threonine-protein kinase
(EC:2.7.11.1); K08827 serine/threonine-protein kinase PRP4
[EC:2.7.11.1]
Length=895
Score = 211 bits (538), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 94/146 (64%), Positives = 123/146 (84%), Gaps = 0/146 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG +YD +ID+WSAAAT++ELATG +LFPGRTNN ML+ +MEYKGK PN++IR
Sbjct 738 YRAPEIILGLRYDCKIDIWSAAATIFELATGDILFPGRTNNHMLKLMMEYKGKVPNRVIR 797
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
AG LSS HF ++LDFI+ +DSFS +D+ +++ DLR +SITD LLE+Q W+KG+SPK +
Sbjct 798 AGQLSSQHFDDNLDFIYVSRDSFSHKDSVKLVSDLRAKRSITDVLLERQPWIKGTSPKKD 857
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
+ R+MRQLGDLLE+CL +DP KRL+
Sbjct 858 AMVRRMRQLGDLLERCLAIDPAKRLS 883
> pfa:PF11_0156 Ser/Thr protein kinase; K08827 serine/threonine-protein
kinase PRP4 [EC:2.7.11.1]
Length=699
Score = 206 bits (523), Expect = 3e-53, Method: Composition-based stats.
Identities = 93/146 (63%), Positives = 122/146 (83%), Gaps = 0/146 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG +YD+QIDVWSAAAT++ELATG++LFPG++NN M++ +MEYKGK +K+I+
Sbjct 532 YRAPEIILGFRYDAQIDVWSAAATVFELATGKILFPGKSNNHMIKLMMEYKGKFSHKMIK 591
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G S HF E+LDF++ +D +SK++ R++ DLRPTK+IT LLE Q WLKG+SPKM
Sbjct 592 GGQFYSQHFNENLDFLYVDRDHYSKKEVVRVISDLRPTKNITCDLLEHQYWLKGNSPKMQ 651
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
FL++K++QLGDLLEKCLILDP KR T
Sbjct 652 FLKKKIKQLGDLLEKCLILDPSKRYT 677
> tpv:TP01_0303 hypothetical protein; K08827 serine/threonine-protein
kinase PRP4 [EC:2.7.11.1]
Length=796
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 90/146 (61%), Positives = 115/146 (78%), Gaps = 0/146 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG +Y+ +IDVWSAAAT+YELATG +LFPGR NN ML+ +ME+KGK P+K+IR
Sbjct 639 YRAPEIILGCRYNCKIDVWSAAATIYELATGDILFPGRNNNHMLKLMMEFKGKVPSKMIR 698
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
AG SS HF E+LDF++ D +K TR+L DLRPT++ITD + E+Q W K +SPK
Sbjct 699 AGQFSSQHFDENLDFVYTSTDPLTKTTVTRVLQDLRPTRNITDCIFERQPWTKSNSPKKE 758
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
L +K+RQLG+LLEKCL+LDP KR +
Sbjct 759 ILVKKIRQLGELLEKCLVLDPNKRFS 784
> hsa:8899 PRPF4B, KIAA0536, PR4H, PRP4, PRP4H, PRP4K, dJ1013A10.1;
PRP4 pre-mRNA processing factor 4 homolog B (yeast) (EC:2.7.11.1);
K08827 serine/threonine-protein kinase PRP4 [EC:2.7.11.1]
Length=1007
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 64/146 (43%), Positives = 94/146 (64%), Gaps = 7/146 (4%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEII+G YD ID+WS TLYEL TG++LFPG+TNN ML+ M+ KGK PNK+IR
Sbjct 855 YRAPEIIIGKSYDYGIDMWSVGCTLYELYTGKILFPGKTNNHMLKLAMDLKGKMPNKMIR 914
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G+ HF ++L+F++ D ++R+ ++ + PTK + L+ Q +
Sbjct 915 KGVFKDQHFDQNLNFMYIEVDKVTEREKVTVMSTINPTKDLLADLIGCQRLPEDQ----- 969
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
R+K+ QL DLL++ L+LDP KR++
Sbjct 970 --RKKVHQLKDLLDQILMLDPAKRIS 993
> mmu:19134 Prpf4b, 2610037H07, MGC5960, Prp4, Prp4k, Prpk, cbp143;
PRP4 pre-mRNA processing factor 4 homolog B (yeast) (EC:2.7.11.1);
K08827 serine/threonine-protein kinase PRP4 [EC:2.7.11.1]
Length=1007
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 64/146 (43%), Positives = 94/146 (64%), Gaps = 7/146 (4%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEII+G YD ID+WS TLYEL TG++LFPG+TNN ML+ M+ KGK PNK+IR
Sbjct 855 YRAPEIIIGKSYDYGIDMWSVGCTLYELYTGKILFPGKTNNHMLKLAMDLKGKMPNKMIR 914
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G+ HF ++L+F++ D ++R+ ++ + PTK + L+ Q +
Sbjct 915 KGVFKDQHFDQNLNFMYIEVDKVTEREKVTVMSTINPTKDLLADLIGCQRLPEDQ----- 969
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
R+K+ QL DLL++ L+LDP KR++
Sbjct 970 --RKKVHQLKDLLDQILMLDPAKRIS 993
> xla:495151 prpf4b; PRP4 pre-mRNA processing factor 4 homolog
B (EC:2.7.11.1); K08827 serine/threonine-protein kinase PRP4
[EC:2.7.11.1]
Length=991
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 62/146 (42%), Positives = 92/146 (63%), Gaps = 7/146 (4%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEII+G YD ID+WS TLYEL G++LFPG+TNN ML+ M+ KGK PNK+IR
Sbjct 839 YRAPEIIIGKMYDYGIDMWSVGCTLYELYNGKILFPGKTNNHMLKLAMDLKGKMPNKMIR 898
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G+ HF +L+F++ D ++R+ ++ + PTK + L+ Q +
Sbjct 899 KGVFKDQHFDANLNFMYIEVDKVTEREKVTVMSTINPTKDLLTDLIGCQRLPEDQ----- 953
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
R+K+ Q+ DLL++ L+LDP KR++
Sbjct 954 --RKKVHQIKDLLDQILMLDPAKRIS 977
> dre:406758 prpf4b, wu:fc18e10, wu:fe47g02, zgc:55623; PRP4 pre-mRNA
processing factor 4 homolog B (EC:2.7.11.1); K08827
serine/threonine-protein kinase PRP4 [EC:2.7.11.1]
Length=1010
Score = 126 bits (316), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 61/146 (41%), Positives = 92/146 (63%), Gaps = 7/146 (4%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEI++G YD ID+WS TLYEL TG++LFPG +NN ML+ M+ KGK PNK+IR
Sbjct 858 YRAPEIVIGKPYDYGIDMWSVGCTLYELYTGKILFPGSSNNHMLKLAMDVKGKMPNKMIR 917
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G+ HF ++L+F++ D ++R+ ++ + PTK + ++ Q +
Sbjct 918 KGLFKDQHFDQNLNFLYIEVDKVTEREKVTVMSTINPTKDLLADMVGGQRLPEDQ----- 972
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
R+K+ QL DLL+ L+LDP KR++
Sbjct 973 --RKKVMQLKDLLDGTLMLDPAKRIS 996
> cel:F22D6.5 prpf-4; vertebrate Pre-mRNA Processing Factor family
member (prpf-4); K08827 serine/threonine-protein kinase
PRP4 [EC:2.7.11.1]
Length=775
Score = 118 bits (295), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 58/146 (39%), Positives = 87/146 (59%), Gaps = 7/146 (4%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEI+LG ++D ID+WS A TLYE+ TG+++FPGRTNN ML+ + KGK PNKL+R
Sbjct 622 YRAPEIMLGVRHDYAIDLWSVAVTLYEVYTGKIMFPGRTNNHMLKLFTDVKGKYPNKLVR 681
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
HF + + +++ D ++R+ +L +L+PT+ + L+ Q + ++
Sbjct 682 KSQFKDTHFDVNCNLLYQEVDKVTERNKITVLANLKPTRDLESELIAGQRLSRDQMDQIQ 741
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
R LL+ LILDP KR T
Sbjct 742 AFRT-------LLDGMLILDPSKRTT 760
> ath:AT1G13350 protein kinase family protein
Length=761
Score = 114 bits (286), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 61/146 (41%), Positives = 88/146 (60%), Gaps = 10/146 (6%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG YD +D+WS LYEL +G+++FPG TNN+MLR ME KG P K++R
Sbjct 613 YRAPEIILGLPYDHPLDIWSVGCCLYELFSGKIMFPGSTNNEMLRLHMELKGAFPKKMLR 672
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G HF +DL F +DS +++ T R++ +++P + + + KQ + S +
Sbjct 673 KGAFIDQHFDKDLCFYATEEDSVTRKTTKRMMVNIKPKEFGS---VIKQRYKDEDSKLLV 729
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
R DLL++ ILDPQKR+T
Sbjct 730 HFR-------DLLDRIFILDPQKRIT 748
> ath:AT3G25840 protein kinase family protein; K08827 serine/threonine-protein
kinase PRP4 [EC:2.7.11.1]
Length=673
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 62/146 (42%), Positives = 86/146 (58%), Gaps = 11/146 (7%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PEIILG YD +D+WS LYEL +G+VLFPG TNNDMLR ME KG P K++R
Sbjct 526 YRSPEIILGLTYDHPLDIWSVGCCLYELYSGKVLFPGATNNDMLRLHMELKGPFPKKMLR 585
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G HF DL+F +D+ S + R++ +++P S+++ G PK+
Sbjct 586 KGAFIDQHFDHDLNFYATEEDTVSGKLIKRMIVNVKPKD--FGSIIK---GYPGEDPKI- 639
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
+ DLL+K ILDP++RLT
Sbjct 640 -----LAHFRDLLDKMFILDPERRLT 660
> ath:AT3G53640 protein kinase family protein
Length=642
Score = 111 bits (277), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 61/146 (41%), Positives = 86/146 (58%), Gaps = 10/146 (6%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG YD +D+WS LYEL +G+++FPG TNNDMLR ME KG P K++R
Sbjct 494 YRAPEIILGLPYDHPLDIWSVGCCLYELYSGKIMFPGSTNNDMLRLHMELKGPFPKKMLR 553
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G HF +DL F +DS + + RI+ +++P S++ ++ + PK+
Sbjct 554 KGAFIDQHFDKDLCFYATEEDSVTGKTIRRIMVNVKPKD--LGSVIRRR--YEDEDPKV- 608
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
+ +LL+K LDPQKRLT
Sbjct 609 -----LVHFRNLLDKIFTLDPQKRLT 629
> xla:100037169 hypothetical protein LOC100037169
Length=959
Score = 111 bits (277), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 49/106 (46%), Positives = 71/106 (66%), Gaps = 0/106 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEII+G YD ID+WS TLYEL G++LFPG+TNN ML+ M+ KGK PNK+IR
Sbjct 838 YRAPEIIIGKMYDYGIDMWSVGCTLYELYNGKILFPGKTNNHMLKLAMDLKGKMPNKMIR 897
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLL 106
G+ HF +L+F++ D ++R+ ++ + PTK ++ L+
Sbjct 898 KGVCKDQHFDANLNFMYIEVDKVTEREKVTVMSTINPTKDLSADLV 943
> cpv:cgd8_5180 hypothetical protein
Length=454
Score = 107 bits (268), Expect = 9e-24, Method: Composition-based stats.
Identities = 58/149 (38%), Positives = 90/149 (60%), Gaps = 11/149 (7%)
Query 1 YRAPEIILGA-KYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLI 59
YRAPEIILG Y ID+WS LYE TG ++F GRTNNDM++ IMEY+G P +L+
Sbjct 301 YRAPEIILGCISYGQPIDIWSIGCVLYECFTGDIMFKGRTNNDMIKLIMEYRGNFPKRLL 360
Query 60 RAGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLE--KQTWLKGSSP 117
G+ + HF+E L + + + +I+ + K+I ++L++ K + + S
Sbjct 361 NQGVFTKSHFSECLTQFKWI----NTQGMLQIIKNYTANKNIYNNLMDSVKSSNIVYSET 416
Query 118 KMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ +RR L +L+EKCLI+DP+KR++
Sbjct 417 EQTLIRR----LSNLIEKCLIIDPKKRIS 441
> hsa:8444 DYRK3, DYRK5, RED, REDK, hYAK3-2; dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase 3 (EC:2.7.12.1);
K08825 dual-specificity tyrosine-(Y)-phosphorylation regulated
kinase [EC:2.7.12.1]
Length=568
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 34/59 (57%), Positives = 40/59 (67%), Gaps = 0/59 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLI 59
YRAPEIILG++Y + ID+WS L EL TGQ LFPG D L C+ME G PP KL+
Sbjct 355 YRAPEIILGSRYSTPIDIWSFGCILAELLTGQPLFPGEDEGDQLACMMELLGMPPPKLL 413
> dre:564976 dyrk4; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 4; K08825 dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase [EC:2.7.12.1]
Length=634
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 48/146 (32%), Positives = 66/146 (45%), Gaps = 25/146 (17%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE+ILG Y ID+WS L EL TG LFPG + + + CIME G PPN ++
Sbjct 378 YRSPEVILGHPYSMAIDMWSLGCILAELYTGYPLFPGESEVEQIACIMEIMGLPPNDFVQ 437
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
L F + + KR RP+ S+L+ L
Sbjct 438 TASRRRLFFDSKGNPRNITNSKGKKR---------RPSSKDLASVLKTNDPL-------- 480
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
FL D +++CL+ DP KR+T
Sbjct 481 FL--------DFIQRCLVWDPTKRMT 498
> mmu:226419 Dyrk3, BC006704; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 3 (EC:2.7.12.1); K08825 dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase [EC:2.7.12.1]
Length=586
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 33/59 (55%), Positives = 39/59 (66%), Gaps = 0/59 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLI 59
YRAPEIILG +Y + ID+WS L EL TGQ LFPG D L C++E G PP KL+
Sbjct 374 YRAPEIILGCRYSTPIDIWSFGCILAELLTGQPLFPGEDEGDQLACMIELLGMPPQKLL 432
> dre:100141364 zgc:172180; K08825 dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase [EC:2.7.12.1]
Length=581
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 50/149 (33%), Positives = 69/149 (46%), Gaps = 14/149 (9%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILG++Y ID+WS L EL TG LFPG D L C+ME G PP+KL+
Sbjct 366 YRAPEVILGSRYGMPIDMWSFGCILAELLTGYPLFPGEDEGDQLACMMELLGMPPSKLLE 425
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTW---LKGSSP 117
+ + + + S + T +L+ R + + W LKG
Sbjct 426 QAKRAKNFISSKGHPRYCTVSTLS--NGTIVLNGSRSRRGKMRGAPGSKEWGTALKGCE- 482
Query 118 KMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
F+ D L+KCL DP R+T
Sbjct 483 DATFI--------DFLKKCLDWDPSTRMT 503
> xla:496058 dyrk3; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 3; K08825 dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase [EC:2.7.12.1]
Length=567
Score = 72.0 bits (175), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 32/59 (54%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLI 59
YRAPE+ILG++Y ID+WS L EL TG LFPG D L C+ME G PP KL+
Sbjct 355 YRAPEVILGSRYGMPIDIWSFGCILVELLTGYPLFPGEDEGDQLACMMELLGAPPPKLL 413
> mmu:69181 Dyrk2, 1810038L18Rik; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 2 (EC:2.7.12.1); K08825 dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase
[EC:2.7.12.1]
Length=599
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 51/149 (34%), Positives = 70/149 (46%), Gaps = 14/149 (9%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILGA+Y ID+WS L EL TG L PG D L C++E G P KL+
Sbjct 386 YRAPEVILGARYGMPIDMWSLGCILAELLTGYPLLPGEDEGDQLACMIELLGMPSQKLLD 445
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTW---LKGSSP 117
A + +F + R + D + +L+ R + E + W LKG
Sbjct 446 ASKRAK-NFVSSKGYP-RYCTVTTLSDGSVVLNGGRSRRGKLRGPPESREWGNALKGCDD 503
Query 118 KMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ FL D L++CL DP R+T
Sbjct 504 PL-FL--------DFLKQCLEWDPAVRMT 523
> hsa:8445 DYRK2, FLJ21217, FLJ21365; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 2 (EC:2.7.12.1); K08825
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase
[EC:2.7.12.1]
Length=528
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 51/149 (34%), Positives = 70/149 (46%), Gaps = 14/149 (9%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILGA+Y ID+WS L EL TG L PG D L C++E G P KL+
Sbjct 315 YRAPEVILGARYGMPIDMWSLGCILAELLTGYPLLPGEDEGDQLACMIELLGMPSQKLLD 374
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTW---LKGSSP 117
A + +F + R + D + +L+ R + E + W LKG
Sbjct 375 ASKRAK-NFVSSKGYP-RYCTVTTLSDGSVVLNGGRSRRGKLRGPPESREWGNALKGCDD 432
Query 118 KMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ FL D L++CL DP R+T
Sbjct 433 PL-FL--------DFLKQCLEWDPAVRMT 452
> cel:F49E11.1 mbk-2; MiniBrain Kinase (Drosophila) homolog family
member (mbk-2); K08825 dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase [EC:2.7.12.1]
Length=796
Score = 68.2 bits (165), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 66/160 (41%), Gaps = 36/160 (22%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILG KY ID+WS L EL TG L PG ND L I+E G PP K +
Sbjct 627 YRAPEVILGTKYGMPIDMWSLGCILAELLTGYPLLPGEDENDQLALIIELLGMPPPKSLE 686
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
+KR T I P S+ + L G+ K
Sbjct 687 T----------------------AKRARTFITSKGYPRYCTATSMPDGSVVLAGARSKRG 724
Query 121 FLR---------RKMRQLG-----DLLEKCLILDPQKRLT 146
+R ++ +G D L++CL DP+ R+T
Sbjct 725 KMRGPPASRSWSTALKNMGDELFVDFLKRCLDWDPETRMT 764
> mmu:101320 Dyrk4, AW049118, Dyrk4a, Dyrk4b; dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase 4 (EC:2.7.12.1);
K08825 dual-specificity tyrosine-(Y)-phosphorylation regulated
kinase [EC:2.7.12.1]
Length=632
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 47/146 (32%), Positives = 67/146 (45%), Gaps = 25/146 (17%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE+ILG Y+ ID+WS + EL TG LFPG + L CIME G PP
Sbjct 385 YRSPEVILGHPYNMAIDMWSLGCIMAELYTGYPLFPGENEVEQLACIMEVLGLPP----- 439
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
HFT+ R + F + + +++ R K DS K + + +
Sbjct 440 ------AHFTQTAS---RRQVFFDSKGLPKNINNNRGGKRYPDS---KDLTMVVKTYDSS 487
Query 121 FLRRKMRQLGDLLEKCLILDPQKRLT 146
FL D L +CL+ +P R+T
Sbjct 488 FL--------DFLRRCLVWEPSLRMT 505
> hsa:28996 HIPK2, DKFZp686K02111, FLJ23711, PRO0593; homeodomain
interacting protein kinase 2 (EC:2.7.11.1); K08826 homeodomain
interacting protein kinase [EC:2.7.11.1]
Length=1171
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 49/161 (30%), Positives = 72/161 (44%), Gaps = 25/161 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 367 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 426
Query 61 AGMLSSLHFTEDLDFIHR---------------VKDSFSKRDTTRILHDLRPTKSITDSL 105
AG ++ F D D + +K +++ L D+ TD
Sbjct 427 AGTKTTRFFNRDTDSPYPLWRLKTPDDHEAETGIKSKEARKYIFNCLDDMAQVNMTTD-- 484
Query 106 LEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
L+GS M + R+ DLL+K L +D KR+T
Sbjct 485 ------LEGSD--MLVEKADRREFIDLLKKMLTIDADKRIT 517
> dre:557602 dyrk2, fc18c09, si:ch211-266a5.8, wu:fa09g03, wu:fc18c09;
dual-specificity tyrosine-(Y)-phosphorylation regulated
kinase 2 (EC:2.7.12.1); K08825 dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase [EC:2.7.12.1]
Length=587
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 50/154 (32%), Positives = 70/154 (45%), Gaps = 24/154 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILGA+Y ID+WS L EL TG L PG D L C++E G P KL+
Sbjct 375 YRAPEVILGARYGMPIDMWSLGCILAELLTGYPLLPGEDEGDQLACVIEMLGMPSEKLLE 434
Query 61 AGMLSSLHFTEDLDFIH-----RVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTW---L 112
+ + +FI R + D + +L+ R + + W L
Sbjct 435 SSKRAK-------NFISSKGHPRYCTVTTLPDASVVLNGGRSRRGKLRGPPGSRDWVTAL 487
Query 113 KGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
KG + FL D L++CL DP +R+T
Sbjct 488 KGCDDPL-FL--------DFLKQCLEWDPSQRMT 512
> mmu:15258 Hipk2, 1110014O20Rik, B230339E18Rik, Stank; homeodomain
interacting protein kinase 2 (EC:2.7.11.1); K08826 homeodomain
interacting protein kinase [EC:2.7.11.1]
Length=1196
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 49/161 (30%), Positives = 72/161 (44%), Gaps = 25/161 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 367 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 426
Query 61 AGMLSSLHFTEDLDFIHR---------------VKDSFSKRDTTRILHDLRPTKSITDSL 105
AG ++ F D D + +K +++ L D+ TD
Sbjct 427 AGTKTTRFFNRDTDSPYPLWRLKTPDDHEAETGIKSKEARKYIFNCLDDMAQVNMTTD-- 484
Query 106 LEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
L+GS M + R+ DLL+K L +D KR+T
Sbjct 485 ------LEGSD--MLVEKADRREFIDLLKKMLTIDADKRVT 517
> hsa:8798 DYRK4; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 4 (EC:2.7.12.1); K08825 dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase [EC:2.7.12.1]
Length=520
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/62 (48%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE+ILG YD ID+WS EL TG LFPG + L CIME G PP I+
Sbjct 270 YRSPEVILGHPYDVAIDMWSLGCITAELYTGYPLFPGENEVEQLACIMEVLGLPPAGFIQ 329
Query 61 AG 62
Sbjct 330 TA 331
> xla:100189543 hipk1; homeodomain interacting protein kinase
1 (EC:2.7.11.1)
Length=1173
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 49/161 (30%), Positives = 71/161 (44%), Gaps = 25/161 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 356 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 415
Query 61 AGMLSSLHFTEDLDF-----------IHRVKDSFSKRDTTR----ILHDLRPTKSITDSL 105
AG +S F D D H V+ ++ + L D+ TD
Sbjct 416 AGTKTSRFFNRDPDLGYPLWRLKAPDEHEVETGIKSKEARKYIFNCLDDMAQVNMSTD-- 473
Query 106 LEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
LE L + + ++ DLL+K L +D KR+T
Sbjct 474 LEGTDMLAEKADRREYI--------DLLKKMLTIDADKRIT 506
> dre:557551 hipk2, MGC198433, cb1009, si:dkey-16j24.2, si:dkey-239k7.1,
wu:fk56d12; homeodomain interacting protein kinase
2 (EC:2.7.11.1); K08826 homeodomain interacting protein kinase
[EC:2.7.11.1]
Length=1226
Score = 65.9 bits (159), Expect = 4e-11, Method: Composition-based stats.
Identities = 49/161 (30%), Positives = 71/161 (44%), Gaps = 25/161 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 381 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 440
Query 61 AGMLSSLHFTEDLDFIHR---------------VKDSFSKRDTTRILHDLRPTKSITDSL 105
AG ++ F D D + +K +++ L D+ TD
Sbjct 441 AGTKTTRFFNRDPDSTYPLWRLKTPEDHEAETGIKSKEARKYIFNCLDDMAQVNMTTD-- 498
Query 106 LEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
L+GS M + R+ DLL K L +D KR+T
Sbjct 499 ------LEGSD--MLAEKADRREFIDLLTKMLTIDADKRIT 531
> cel:T04C10.1 mbk-1; MiniBrain Kinase (Drosophila) homolog family
member (mbk-1); K08825 dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase [EC:2.7.12.1]
Length=882
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 42/149 (28%), Positives = 74/149 (49%), Gaps = 8/149 (5%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE++LG YD++ID+WS L E+ TG+ LF G + D + I+E G PP +++
Sbjct 496 YRSPEVLLGIAYDTKIDMWSLGCILVEMHTGEPLFAGSSEVDQMMKIVEVLGMPPKEMLD 555
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKG---SSP 117
G + +F + D I+ K + RD R + + + + L G P
Sbjct 556 IGPKTHKYFDKTEDGIYYCKKT---RDGYRHTYKAPGARKLHEILGVTSGGPGGRRLGEP 612
Query 118 KMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ + DL+++ L DP++R++
Sbjct 613 GHSV--EDYSKFKDLIKRMLQFDPKQRIS 639
> xla:735128 MGC115587; serine/threonine-protein kinase SRPK1-like
Length=386
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 48/82 (58%), Gaps = 6/82 (7%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLF---PGRT---NNDMLRCIMEYKGKP 54
YRAPE++LG+ Y + +D+WS A +E+AT LF G+T +D + CIME G+
Sbjct 248 YRAPEVLLGSTYSTSVDIWSTACMAFEMATSYYLFEPHAGKTFTREDDHIACIMELLGRI 307
Query 55 PNKLIRAGMLSSLHFTEDLDFI 76
P K+I +G S F + D +
Sbjct 308 PPKVISSGRKSPAFFNKQGDLL 329
> dre:572323 homeodomain-interacting protein kinase 3-like; K08826
homeodomain interacting protein kinase [EC:2.7.11.1]
Length=1152
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 51/151 (33%), Positives = 74/151 (49%), Gaps = 7/151 (4%)
Query 2 RAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIRA 61
RAPEIILG + ID+WS + EL G L+PG + D +R I + +G PP L+ A
Sbjct 363 RAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPPEYLLSA 422
Query 62 GMLSSLHFTEDLDFIH---RVKDSFSKRDTTRILHDLRPTKSITDSLLE-KQTWLKGSSP 117
G +S F D + R+K S+ ++ + K I + L + Q L
Sbjct 423 GTKTSRFFNRGPDSSYPLWRLKTP-SEHESEMGIKSKEARKYIFNCLDDMMQVNLPNHLE 481
Query 118 KMNFLRRKM--RQLGDLLEKCLILDPQKRLT 146
+ L K R+L DLL++ L LD KR+T
Sbjct 482 GTDLLAEKADRRELIDLLKRMLRLDADKRIT 512
> tgo:TGME49_083480 dual-specificity tyrosine-phosphorylation-regulated
kinase, putative (EC:2.7.12.1); K08825 dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase [EC:2.7.12.1]
Length=956
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 36/98 (36%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE++LG Y ID+WS L EL TG LF G D + CIME G PP +I
Sbjct 795 YRSPEVLLGLSYGCPIDIWSLGCILAELQTGHPLFAGENETDQVSCIMEILGPPPLYIIA 854
Query 61 AGMLSSLHFTEDLD-----FIHRVKDSFSKRDTTRILH 93
+ F + D H K S RD + +L
Sbjct 855 KSPRRRIFFDSNGDPKPYTNSHGKKRRVSSRDLSTVLQ 892
> xla:495423 hipk1; homeodomain interacting protein kinase 1 (EC:2.7.11.1);
K08826 homeodomain interacting protein kinase
[EC:2.7.11.1]
Length=1173
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 47/161 (29%), Positives = 70/161 (43%), Gaps = 25/161 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 358 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 417
Query 61 AGMLSSLHFTEDLDFIHR---------------VKDSFSKRDTTRILHDLRPTKSITDSL 105
AG +S F + D + +K +++ L D+ TD
Sbjct 418 AGTKTSRFFNRNPDLGYSLWRLKAPDEHEMETGIKSKEARKYIFNCLDDMAQVNMSTD-- 475
Query 106 LEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
LE L + + ++ DLL K L +D KR+T
Sbjct 476 LEGTDMLAEKADRREYI--------DLLNKMLTIDADKRIT 508
> xla:495265 hipk2; homeodomain interacting protein kinase 2 (EC:2.7.11.1);
K08826 homeodomain interacting protein kinase
[EC:2.7.11.1]
Length=1154
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 50/161 (31%), Positives = 73/161 (45%), Gaps = 25/161 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 356 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 415
Query 61 AGMLSSLHFTEDLDF---IHRVKDSFSKRDTTRI------------LHDLRPTKSITDSL 105
+G ++ F D D + R+K + T I L D+ +D
Sbjct 416 SGTKTTRFFNRDGDSPYPLWRLKTPEDHENETGIKSKEARKYIFNCLDDMAQVNMTSD-- 473
Query 106 LEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
L+GS + R R+ DLL+K L +D KR+T
Sbjct 474 ------LEGSDLLVEKADR--REFIDLLKKMLTIDADKRIT 506
> mmu:15259 Hipk3, DYRK6, FIST3, PKY; homeodomain interacting
protein kinase 3 (EC:2.7.11.1); K08826 homeodomain interacting
protein kinase [EC:2.7.11.1]
Length=1191
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 45/152 (29%), Positives = 72/152 (47%), Gaps = 7/152 (4%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG D +R I + +G P +L+
Sbjct 365 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGALEYDQIRYISQTQGLPGEQLLN 424
Query 61 AGMLSSLHFTEDLDFIH---RVKDSFSKRDTTRILHDLRPTKSITDSL---LEKQTWLKG 114
G S+ F + D H R+K + + + + K I +SL + T +
Sbjct 425 VGTKSTRFFCRETDMSHSGWRLK-TLEEHEAETGMKSKEARKYIFNSLDDIVHVNTVMDL 483
Query 115 SSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ + R+ +LL+K L++D R+T
Sbjct 484 EGGDLLAEKADRREFVNLLKKMLLIDADLRIT 515
> ath:AT2G40120 protein kinase family protein
Length=570
Score = 63.2 bits (152), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/82 (37%), Positives = 47/82 (57%), Gaps = 0/82 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILG YD +ID+WS L EL +G+VLFP +L I+ G +++
Sbjct 431 YRAPEVILGLPYDEKIDLWSLGCILAELCSGEVLFPNEAVAMILARIVAVLGPIETEMLE 490
Query 61 AGMLSSLHFTEDLDFIHRVKDS 82
G + +FT++ D H ++S
Sbjct 491 KGQETHKYFTKEYDLYHLNEES 512
> hsa:10114 HIPK3, DYRK6, FIST3, PKY, YAK1; homeodomain interacting
protein kinase 3 (EC:2.7.11.1); K08826 homeodomain interacting
protein kinase [EC:2.7.11.1]
Length=1194
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 49/156 (31%), Positives = 75/156 (48%), Gaps = 15/156 (9%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG D +R I + +G P +L+
Sbjct 365 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGALEYDQIRYISQTQGLPGEQLLN 424
Query 61 AGMLSSLHFTEDLDFIH---RVKDSFSKRDTTRILHDLRPTKSITDSL-----LEKQTWL 112
G S+ F ++ D H R+K + + + + K I +SL + L
Sbjct 425 VGTKSTRFFCKETDMSHSGWRLK-TLEEHEAETGMKSKEARKYIFNSLDDVAHVNTVMDL 483
Query 113 KGSSPKMNFLRRKM--RQLGDLLEKCLILDPQKRLT 146
+GS + L K R+ LL+K L++D R+T
Sbjct 484 EGS----DLLAEKADRREFVSLLKKMLLIDADLRIT 515
> dre:556212 dyrk1b, si:ch211-225p5.1; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 1B (EC:2.7.12.1); K08825
dual-specificity tyrosine-(Y)-phosphorylation regulated
kinase [EC:2.7.12.1]
Length=681
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 48/158 (30%), Positives = 76/158 (48%), Gaps = 20/158 (12%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE++LG YD ID+WS L E+ TG+ LF G D + I+E G PPN ++
Sbjct 279 YRSPEVLLGMPYDLAIDMWSLGCILVEMHTGEPLFSGSNEVDQMNKIVEVLGVPPNHMLD 338
Query 61 AGMLSSLHFTEDLDFIHRVKD------------SFSKRDTTRILHDLRPTKSITDSLLEK 108
+ +F + D + VK S K TR LH++ + T +
Sbjct 339 QAPKARKYFDKLSDGLWTVKKNKDIKKVLYPSASEYKPPATRRLHEILGVE--TGGPGGR 396
Query 109 QTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ +G +P ++L+ K DL+ + L DP+ R+T
Sbjct 397 RAGEQGHAP-CDYLKFK-----DLILRMLDYDPKTRIT 428
> hsa:204851 HIPK1, KIAA0630, MGC26642, MGC33446, MGC33548, Myak,
Nbak2; homeodomain interacting protein kinase 1 (EC:2.7.11.1);
K08826 homeodomain interacting protein kinase [EC:2.7.11.1]
Length=1075
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 50/154 (32%), Positives = 73/154 (47%), Gaps = 11/154 (7%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 358 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 417
Query 61 AGMLSSLHFTEDLDF---IHRVKDSFSKRDTTRILHDLRPTKSITDSL-----LEKQTWL 112
AG ++ F D + + R+K T I K I + L + T L
Sbjct 418 AGTKTTRFFNRDPNLGYPLWRLKTPEEHELETGI-KSKEARKYIFNCLDDMAQVNMSTDL 476
Query 113 KGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+G+ M + R+ DLL+K L +D KR+T
Sbjct 477 EGTD--MLAEKADRREYIDLLKKMLTIDADKRIT 508
> xla:379954 dyrk1a.2, MGC52802; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 1A, gene 2 (EC:2.7.12.1);
K08825 dual-specificity tyrosine-(Y)-phosphorylation regulated
kinase [EC:2.7.12.1]
Length=671
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 48/151 (31%), Positives = 77/151 (50%), Gaps = 13/151 (8%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YR+PE++LG YD ID+WS L E+ TG+ LF G D + I+E G PPN ++
Sbjct 279 YRSPEVLLGMPYDLAIDMWSLGCILVEMHTGEPLFSGSNEVDQMNKIVEVLGTPPNHMLD 338
Query 61 AGMLSSLHFTEDLDFIHRVKDSFS-KRD----TTRILHDLRPTKSITDSLLEKQTWLKGS 115
+ +F + + VK + K+D TR LH++ + T ++ +G
Sbjct 339 QAPKARKYFDKLPEGTWTVKKNKDLKKDYKVPGTRRLHEVLGVE--TGGPGGRRGGEQGH 396
Query 116 SPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
SP ++L+ K DL+ + L DP+ R+T
Sbjct 397 SPS-DYLKFK-----DLILRMLDYDPKTRIT 421
> mmu:15257 Hipk1, 1110062K04Rik, Myak; homeodomain interacting
protein kinase 1 (EC:2.7.11.1); K08826 homeodomain interacting
protein kinase [EC:2.7.11.1]
Length=1210
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 50/154 (32%), Positives = 73/154 (47%), Gaps = 11/154 (7%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 358 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 417
Query 61 AGMLSSLHFTEDLDF---IHRVKDSFSKRDTTRILHDLRPTKSITDSL-----LEKQTWL 112
AG ++ F D + + R+K T I K I + L + T L
Sbjct 418 AGTKTTRFFNRDPNLGYPLWRLKTPEEHELETGI-KSKEARKYIFNCLDDMAQVNMSTDL 476
Query 113 KGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+G+ M + R+ DLL+K L +D KR+T
Sbjct 477 EGTD--MLAEKADRREYIDLLKKMLTIDADKRIT 508
> dre:572042 homeodomain-interacting protein kinase 3-like; K08826
homeodomain interacting protein kinase [EC:2.7.11.1]
Length=1139
Score = 62.4 bits (150), Expect = 5e-10, Method: Composition-based stats.
Identities = 51/165 (30%), Positives = 69/165 (41%), Gaps = 34/165 (20%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPEIILG + ID+WS + EL G L+PG + D +R I + +G P L+
Sbjct 369 YRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLS 428
Query 61 AGMLSSLHFTEDLDFIHRVKDSF-------------------SKRDTTRILHDLRPTKSI 101
AG +S F HR DS SK I + L +
Sbjct 429 AGTKTS-------RFFHRGPDSSYPLWRLKTPAEHEAEFGIKSKEARKYIFNCLDDMMQV 481
Query 102 TDSLLEKQTWLKGSSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
+ LE L + + F+ DLL+K L LD KR+T
Sbjct 482 NMTNLEGTDILAEKADRREFI--------DLLKKMLTLDADKRIT 518
> ath:AT4G03175 protein kinase family protein
Length=139
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 47/81 (58%), Gaps = 0/81 (0%)
Query 2 RAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIRA 61
RAPE+ILG YD +ID+WS + EL +G+VLFP +L I+ G ++++
Sbjct 2 RAPEVILGLPYDEKIDLWSLGCIVAELCSGEVLFPNEAVAMILARIVAVLGPIETEMLKK 61
Query 62 GMLSSLHFTEDLDFIHRVKDS 82
G + +FT++ D H ++S
Sbjct 62 GQETHKYFTKEYDLYHLNEES 82
> dre:791187 srpk1b, zgc:158800; serine/arginine-rich protein
specific kinase 1b; K08831 SFRS protein kinase [EC:2.7.11.1]
Length=640
Score = 61.6 bits (148), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 48/152 (31%), Positives = 65/152 (42%), Gaps = 31/152 (20%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLR------CIMEYKGKP 54
YR+ E+++G Y + D+WS A +ELATG LF + D R I+E GK
Sbjct 502 YRSLEVLIGTGYGTPADIWSTACMAFELATGDYLFEPHSGEDYSRDEDHIALIIELLGKI 561
Query 55 PNKLIRAGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKG 114
P KL+ G S FT+ D H K L+P + D L+EK W
Sbjct 562 PRKLVMNGKYSKEFFTKKGDLRHITK--------------LKPW-GLQDVLVEKYEW--- 603
Query 115 SSPKMNFLRRKMRQLGDLLEKCLILDPQKRLT 146
R + + D L L L P+KR T
Sbjct 604 -------HREEAQNFSDFLLPMLDLIPEKRAT 628
> ath:AT1G73460 protein kinase family protein
Length=1169
Score = 61.6 bits (148), Expect = 9e-10, Method: Composition-based stats.
Identities = 43/144 (29%), Positives = 68/144 (47%), Gaps = 23/144 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILG YD +IDVWS L EL TG VLF + +L +M G N+++
Sbjct 1028 YRAPEVILGLPYDKKIDVWSLGCILAELCTGNVLFQNDSPASLLARVMGIVGSFDNEMLT 1087
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G S +FT++ R+L++ + + L+ K+T L+ P +
Sbjct 1088 KGRDSHKYFTKN-----------------RMLYERNQESNRLEYLIPKRTSLRHRLPMGD 1130
Query 121 FLRRKMRQLGDLLEKCLILDPQKR 144
+ D + L ++P+KR
Sbjct 1131 ------QGFTDFVAHLLEINPKKR 1148
> ath:AT1G73450 protein kinase, putative
Length=1152
Score = 61.2 bits (147), Expect = 1e-09, Method: Composition-based stats.
Identities = 43/144 (29%), Positives = 68/144 (47%), Gaps = 23/144 (15%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE+ILG YD +IDVWS L EL TG VLF + +L +M G N+++
Sbjct 1011 YRAPEVILGLPYDKKIDVWSLGCILAELCTGNVLFQNDSPASLLARVMGIVGSFDNEMLT 1070
Query 61 AGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKMN 120
G S +FT++ R+L++ + + L+ K+T L+ P +
Sbjct 1071 KGRDSHKYFTKN-----------------RMLYERNQESNRLEYLIPKRTSLRHRLPMGD 1113
Query 121 FLRRKMRQLGDLLEKCLILDPQKR 144
+ D + L ++P+KR
Sbjct 1114 ------QGFTDFVAHLLEINPKKR 1131
> tpv:TP04_0880 protein kinase (EC:2.7.1.37); K00870 protein kinase
[EC:2.7.1.37]
Length=724
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLIR 60
YRAPE++LG YD+QID+WS L EL G++LFP + ++ ++ G PP +++
Sbjct 459 YRAPEVVLGLPYDTQIDIWSLGCILCELYLGRILFPSDNSATLVASMVSLLGPPPAYMLQ 518
Query 61 AGMLS 65
M S
Sbjct 519 HKMNS 523
> mmu:20815 Srpk1, AU017960; serine/arginine-rich protein specific
kinase 1 (EC:2.7.11.1); K08831 SFRS protein kinase [EC:2.7.11.1]
Length=648
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 46/86 (53%), Gaps = 6/86 (6%)
Query 1 YRAPEIILGAKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLR------CIMEYKGKP 54
YR+ E+++G+ Y++ D+WS A +ELATG LF + D R I+E GK
Sbjct 510 YRSLEVLIGSGYNTPADIWSTACMAFELATGDYLFEPHSGEDYTRDEDHIALIIELLGKV 569
Query 55 PNKLIRAGMLSSLHFTEDLDFIHRVK 80
P KLI AG S FT+ D H K
Sbjct 570 PRKLIVAGKYSKEFFTKKGDLKHITK 595
> dre:65237 mapk14a, MAPK14, fk28c03, hm:zeh1243, p38, p38a, wu:fk28c03,
zp38a; mitogen-activated protein kinase 14a (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 66/147 (44%), Gaps = 37/147 (25%)
Query 1 YRAPEIILG-AKYDSQIDVWSAAATLYELATGQVLFPGRTNNDMLRCIMEYKGKPPNKLI 59
YRAPEI+L Y+ +D+WS + EL TG+ LFPG + + L+ IM G PP+ LI
Sbjct 189 YRAPEIMLNWMHYNVTVDIWSVGCIMAELLTGRTLFPGTDHINQLQQIMRLTGTPPSSLI 248
Query 60 RAGMLSSLHFTEDLDFIHRVKDSFSKRDTTRILHDLRPTKSITDSLLEKQTWLKGSSPKM 119
S + E +I + P ++ D + G++P
Sbjct 249 -----SRMPSHEARTYISSLPQ--------------MPKRNFADVFI-------GANP-- 280
Query 120 NFLRRKMRQLGDLLEKCLILDPQKRLT 146
Q DLLEK L+LD KR+T
Sbjct 281 --------QAVDLLEKMLVLDTDKRIT 299
Lambda K H
0.321 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40